Reconstructing the last common ancestor of all eukaryotes
- PMID: 39585925
- PMCID: PMC11627563
- DOI: 10.1371/journal.pbio.3002917
Reconstructing the last common ancestor of all eukaryotes
Abstract
Understanding the origin of eukaryotic cells is one of the most difficult problems in all of biology. A key challenge relevant to the question of eukaryogenesis is reconstructing the gene repertoire of the last eukaryotic common ancestor (LECA). As data sets grow, sketching an accurate genomics-informed picture of early eukaryotic cellular complexity requires provision of analytical resources and a commitment to data sharing. Here, we summarise progress towards understanding the biology of LECA and outline a community approach to inferring its wider gene repertoire. Once assembled, a robust LECA gene set will be a useful tool for evaluating alternative hypotheses about the origin of eukaryotes and understanding the evolution of traits in all descendant lineages, with relevance in diverse fields such as cell biology, microbial ecology, biotechnology, agriculture, and medicine. In this Consensus View, we put forth the status quo and an agreed path forward to reconstruct LECA's gene content.
Copyright: © 2024 Richards et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
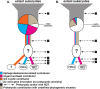
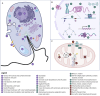

References
-
- Stanier RY, Doudoroff M, Adelberg EA. The microbial world: Prentice-Hall; 1957.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

