Global hinge sites of proteins as target sites for drug binding
- PMID: 39585988
- PMCID: PMC11626116
- DOI: 10.1073/pnas.2414333121
Global hinge sites of proteins as target sites for drug binding
Abstract
Hinge sites of proteins play a key role in mediating conformational mechanics. Among them, those involved in the most collective modes of motion, also called global hinges, are of particular interest, as they support cooperative rearrangements that are often functional. Yet, the utility of targeting global hinges for modulating function remains to be established. We present here a systematic study of a series of proteins resolved in drug-bound forms to examine the probabilistic occurrence of spatial overlaps between hinge sites and drug-binding pockets. Our analysis reveals a high propensity of drug binding to hinge sites compared to random. Notably, one-third of currently approved drugs are colocalized with hinge sites. These mechanosensitive sites are predictable by simple models such as the Gaussian Network Model. Their targeting thus emerges as a viable strategy for developing a new class of drugs that would exploit and modulate the target proteins' intrinsic dynamics, and potentially alleviate drug-resistance when used in combination with orthosteric or allosteric drugs.
Keywords: Gaussian Network Model; collective motions; drug binding sites; ensemble analysis; protein dynamics.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


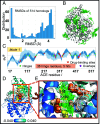


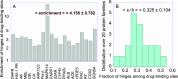
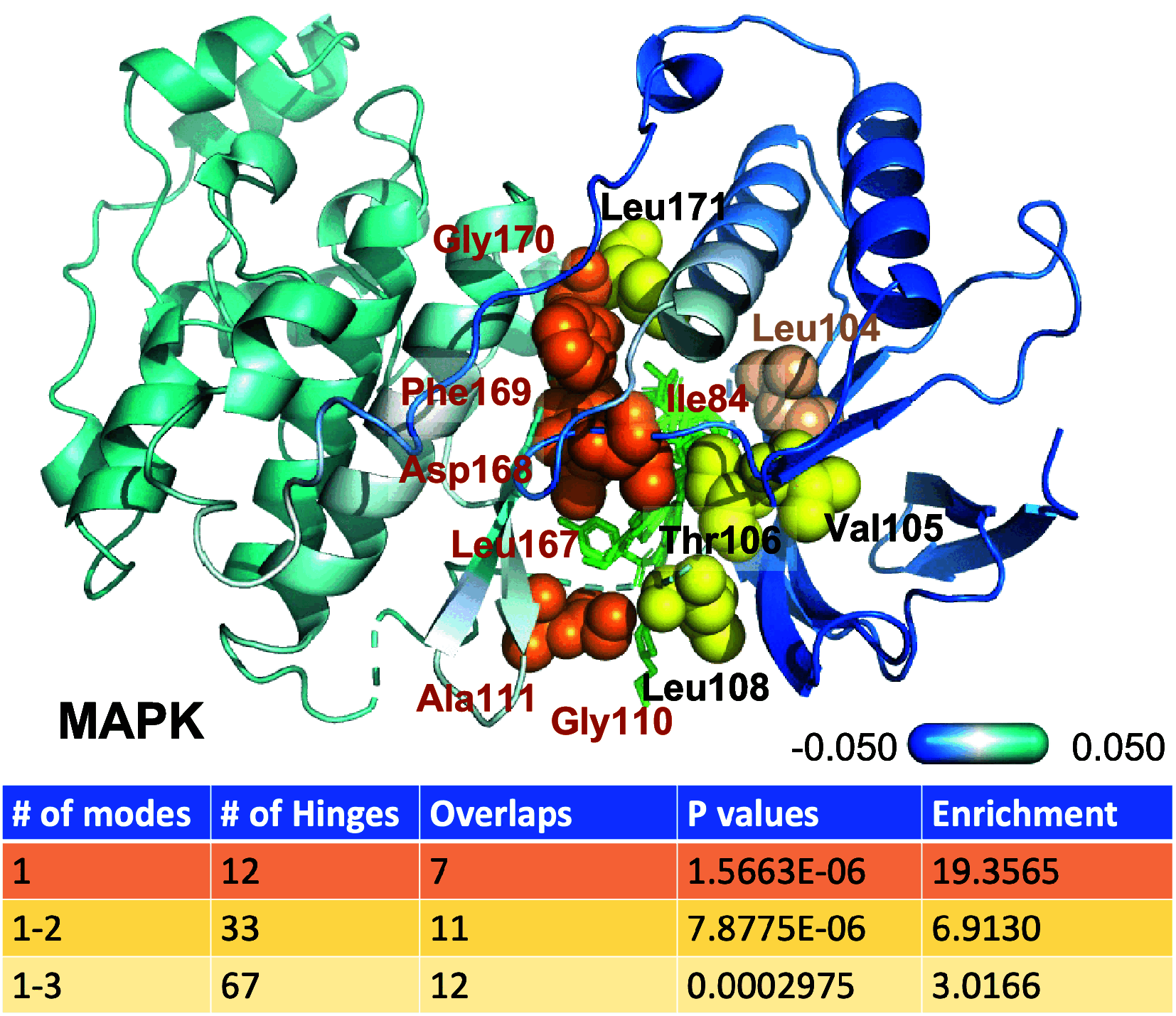
References
-
- Lu S., Shen Q., Zhang J., Allosteric methods and their applications: Facilitating the discovery of allosteric drugs and the investigation of allosteric mechanisms. Acc. Chem. Res. 52, 492–500 (2019). - PubMed
-
- Hardy J. A., Wells J. A., Searching for new allosteric sites in enzymes. Curr. Opin. Struc. Biol. 14, 706–715 (2004). - PubMed
-
- Wells J. A., McClendon C. L., Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450, 1001–1009 (2007). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

