Deep Learning-Based DCE-MRI Automatic Segmentation in Predicting Lesion Nature in BI-RADS Category 4
- PMID: 39586911
- PMCID: PMC12344054
- DOI: 10.1007/s10278-024-01340-2
Deep Learning-Based DCE-MRI Automatic Segmentation in Predicting Lesion Nature in BI-RADS Category 4
Abstract
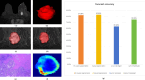
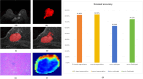
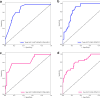
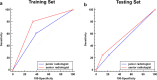
To investigate whether automatic segmentation based on DCE-MRI with a deep learning (DL) algorithm enabled advantages over manual segmentation in differentiating BI-RADS 4 breast lesions. A total of 197 patients with suspicious breast lesions from two medical centers were enrolled in this study. Patients treated at the First Hospital of Qinhuangdao between January 2018 and April 2024 were included as the training set (n = 138). Patients treated at Lanzhou University Second Hospital were assigned to an external validation set (n = 59). Areas of suspicious lesions were delineated based on DL automatic segmentation and manual segmentation, and evaluated consistency through the Dice correlation coefficient. Radiomics models were constructed based on DL and manual segmentations to predict the nature of BI-RADS 4 lesions. Meanwhile, the nature of the lesions was evaluated by both a professional radiologist and a non-professional radiologist. Finally, the area under the curve value (AUC) and accuracy (ACC) were used to determine which prediction model was more effective. Sixty-four malignant cases (32.5%) and 133 benign cases (67.5%) were included in this study. The DL-based automatic segmentation model showed high consistency with manual segmentation, achieving a Dice coefficient of 0.84 ± 0.11. The DL-based radiomics model demonstrated superior predictive performance compared to professional radiologists, with an AUC of 0.85 (95% CI 0.79-0.92). The DL model significantly reduced working time and improved efficiency by 83.2% compared to manual segmentation, further demonstrating its feasibility for clinical applications. The DL-based radiomics model for automatic segmentation outperformed professional radiologists in distinguishing between benign and malignant lesions in BI-RADS category 4, thereby helping to avoid unnecessary biopsies. This groundbreaking progress suggests that the DL model is expected to be widely applied in clinical practice in the near future, providing an effective auxiliary tool for the diagnosis and treatment of breast cancer.
Keywords: BI-RADS 4; Breast cancer; DCE-MRI; Deep learning; Radiomics.
© 2024. The Author(s) under exclusive licence to Society for Imaging Informatics in Medicine.
Conflict of interest statement
Declarations. Ethics Approval: This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of First Hospital of Qinhuangdao (2024K006). Consent to Participate: Informed consent was obtained from all individual participants included in the study. Competing Interests: The authors declare no competing interests.
Figures

References
-
- Siegel R L, Miller K D. Cancer statistics, 2022 [J]. 2022, 72(1): 7–33. - PubMed
-
- SPAK D A, PLAXCO J S, SANTIAGO L, et al. BI-RADS(®) fifth edition: A summary of changes [J]. Diagnostic and interventional imaging, 2017, 98(3): 179-90. - PubMed
-
- GRADISHAR W J, MORAN M S, ABRAHAM J, et al. NCCN Guidelines® Insights: Breast Cancer, Version 4.2021 [J]. Journal of the National Comprehensive Cancer Network : JNCCN, 2021, 19(5): 484-93. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
