HER2-related biomarkers predict clinical outcomes with trastuzumab deruxtecan treatment in patients with HER2-expressing metastatic colorectal cancer: biomarker analyses of DESTINY-CRC01
- PMID: 39587050
- PMCID: PMC11589615
- DOI: 10.1038/s41467-024-53223-3
HER2-related biomarkers predict clinical outcomes with trastuzumab deruxtecan treatment in patients with HER2-expressing metastatic colorectal cancer: biomarker analyses of DESTINY-CRC01
Erratum in
-
Publisher Correction: HER2-related biomarkers predict clinical outcomes with trastuzumab deruxtecan treatment in patients with HER2-expressing metastatic colorectal cancer: biomarker analyses of DESTINY-CRC01.Nat Commun. 2025 Jan 15;16(1):704. doi: 10.1038/s41467-025-56170-9. Nat Commun. 2025. PMID: 39814812 Free PMC article. No abstract available.
Abstract
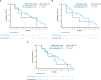
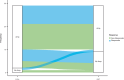
DESTINY-CRC01 (NCT03384940) was a multicentre, open-label, phase 2 study that investigated the safety and efficacy of trastuzumab deruxtecan (T-DXd) in patients with human epidermal growth factor receptor 2 (HER2)-expressing metastatic colorectal cancer (CRC). The present exploratory biomarker analysis aims to investigate relationships between biomarkers and clinical outcomes in patients with HER2-positive (immunohistochemistry [IHC] 3+ or IHC 2+ and in situ hybridization [ISH] positive) Cohort A (N = 53) of DESTINY-CRC01. Higher levels of HER2 biomarkers in baseline tissue and liquid biopsies, including HER2 status (IHC/ISH), HER2/CEP17 ratio, HER2 ISH signals, HER2 H-score, plasma HER2 (ERBB2) amplification status, HER2 adjusted plasma copy number, and HER2 extracellular domain correlate with antitumor activity (indicated by objective response rate, progression-free survival, and overall survival) of T-DXd. Baseline circulating tumor DNA (ctDNA) analysis suggests antitumor activity of T-DXd in patients who had baseline activating RAS, PIK3CA, or HER2 mutations detected in ctDNA.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare the following competing interests: S.S. provided medical writing support to Daiichi Sankyo on this manuscript. In the past 36 months, he received payment for participating on data safety monitoring boards or advisory boards for Agenus, AstraZeneca, Bayer, BMS, CheckmAb, Daiichi Sankyo, GSK, Guardant Health, Merck, Novartis, Pierre-Fabre, Roche Genentech, Seagen, and T-One Therapeutics. K.R. provided research support to Daiichi Sankyo for this study. In the past 36 months, he received grants or contracts from Bayer, Merck, Guardant, Hibercell, Daiichi Sankyo, and Seagen; received payments for educational events from Daiichi Sankyo, Bayer, and Seagen; and received payment for participating on data safety monitoring boards or advisory boards from Seagen and Merck. T.M. received grants or contracts in the past 36 months from Amgen, Boehringer Ingelheim, Cimic Shift Zero, Daiichi Sankyo, Merck, Novartis, ONO Pharmaceutical, Pfizer, Syneos Health Clinical, and Eli Lilly and received payments for educational events from Bayer, Bristol Myers Squibb, Chugai, Daiichi Sankyo, Lilly Japan, Merck BioPharma, ONO Pharmaceutical, Sanofi, Taiho, Takeda, and Yakult Honsha. K.Y. reports support from Daiichi Sankyo for the present manuscript, grants from Taiho Pharmaceutical, and payment or honoraria from Daiichi Sankyo Co. Ltd, Chugai Pharmaceutical Co. Ltd, Bristol Myers Squibb, Eli Lilly Japan, Taiho Pharmaceutical Co. Ltd, Ono Pharmaceutical Co. Ltd, Takeda Pharmaceutical Co. Ltd, and Merck Biopharm Co. Ltd. T.N. received honoraria for lectures from Bristol Myers Squibb, Chugai Pharmaceutical, Daiichi Sankyo Pharmaceutical, Eli Lilly Japan, Merck Serono Pharmaceutical, Ono Pharmaceutical, Takeda Pharmaceutical, Taiho Pharmaceutical, and Yakult-Honsha and was a Data Safety Monitoring Board member for Janssen Pharmaceutical. E.E. received consultancy fees from Amgen, Bayer, Hoffmann-La Roche, Merck Serono, MSD, Novartis, Organon, Pierre Fabre, Sanofi, and Servier; received payments for educational events from Amgen, Bayer, Hoffmann-La Roche, Merck Serono, MSD, Novartis, Organon, Pierre Fabre, Sanofi, and Servier; and received payments for participating on advisory boards from Amgen, Bayer, Hoffmann-La Roche, Merck Serono, MSD, Novartis, Organon, Pierre Fabre, Sanofi, and Servier. J.R. reports no conflicts of interest. I.C. has been on advisory boards for Eli Lilly, Bristol Myers Squibb, MSD, Roche, Merck-Serono, AstraZeneca, OncXerna, Pierre Fabre, Boehringer Ingelheim, Incyte, Astellas, GlaxoSmithKline, Sotio, Eisai, Daiichi Sankyo, Taiho, Servier, Seagen, and Turning Point Therapeutics, reports research funding from Eli Lilly and Janssen Cilag, and honorarium from Eli Lilly, Eisai, Servier, Roche, BMS, and Novartis. M.D.B. reports no conflicts of interest. H.K. received grants or contracts in the past 36 months from Bristol Myers Squibb, Eisai Co. Ltd, Kobayashi Pharmaceutical, and Taiho Pharmaceutical; received consultancy fees from Daiichi Sankyo; received payments for educational events from Bayer Yakuhin, Bristol Myers Squibb, Chugai Pharmaceuticals, Daiichi Sankyo, Eli Lilly Japan, Merck Biopharma, MSD, ONO Pharmaceutical, Otsuka, Taiho Pharmaceutical Takeda, Teijin, and Yakult. F.S. reports no conflicts of interest. M.K. reports no conflicts of interest. K.I. is an employee of Daiichi Sankyo RD Novare, which is a subsidiary of Daiichi Sankyo. Y.K is an employee of and has stock options with Daiichi Sankyo. I.T. reports no conflicts of interest. D.B. reports no conflicts of interest. K.K. reports no conflicts of interest. A.G. reports consulting fees from Daiichi Sankyo, Bayer, Merck/MSD, Genentech/Roche, Natera, and BMS, payment/honoraria from Bayer, Genentech/Roche, and Merck/MSD, and participation on a data safety monitoring board or advisory board for Regeneron. T.Y. received grants or contracts from Amgen K.K., Chugai Pharmaceuticals, Daiichi Sankyo, Genomedia, MSD K.K., Nippon Boehringer Ingelheim, ONO Pharmaceutical, Parexel International, Pfizer Japan, Sanofi K.K., Sysmex, and Taiho and received payments for educational events from Bayer Yakuhin, Chugai, Eli Lilly Japan, Merck Biopharma, MSD, ONO Pharmaceutical, and Taiho.
Figures






References
-
- Sung, H., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021). - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

