Inhibition of lung tumorigenesis by transient reprogramming in cancer cells
- PMID: 39587064
- PMCID: PMC11589828
- DOI: 10.1038/s41419-024-07207-2
Inhibition of lung tumorigenesis by transient reprogramming in cancer cells
Abstract
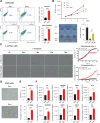
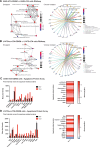
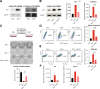
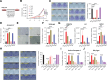
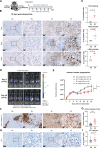
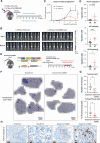
Oncogenic transformation and Oct4, Sox2, Klf4 and c-Myc (OSKM)-mediated induction of pluripotency are two independent and incompatible cellular fates. While continuous expression of OSKM can convert normal somatic cells into teratogenic pluripotent cells, it remains speculative what is the impact of transient OSKM expression in cancer cells. Here, we find that OSKM expression limits the growth of transformed lung cells by inducing apoptosis and senescence. We identify Oct4 and Klf4 as the main individual reprogramming factors responsible for this effect. Mechanistically, the induction of cell cycle inhibitor p21 downstream of the reprogramming factors acts as mediator of cell death and senescence. Using a variety of in vivo systems, including allografts, orthotopic transplantation and KRAS-driven lung cancer mouse models, we demonstrate that transient reprogramming by OSKM expression in cancer cells impairs tumor growth and reduces tumor burden. Altogether, our results show that the induction of transient reprogramming in cancer cells is antitumorigenic opening novel potential therapeutic avenues in oncology.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

