Critical assessment of LC3/GABARAP ligands used for degrader development and ligandability of LC3/GABARAP binding pockets
- PMID: 39587067
- PMCID: PMC11589570
- DOI: 10.1038/s41467-024-54409-5
Critical assessment of LC3/GABARAP ligands used for degrader development and ligandability of LC3/GABARAP binding pockets
Abstract
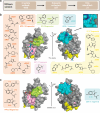
Recent successes in developing small molecule degraders that act through the ubiquitin system have spurred efforts to extend this technology to other mechanisms, including the autophagosomal-lysosomal pathway. Therefore, reports of autophagosome tethering compounds (ATTECs) have received considerable attention from the drug development community. ATTECs are based on the recruitment of targets to LC3/GABARAP, a family of ubiquitin-like proteins that presumably bind to the autophagosome membrane and tether cargo-loaded autophagy receptors into the autophagosome. In this work, we rigorously tested the target engagement of the reported ATTECs to validate the existing LC3/GABARAP ligands. Surprisingly, we were unable to detect interaction with their designated target LC3 using a diversity of biophysical methods. Intrigued by the idea of developing ATTECs, we evaluated the ligandability of LC3/GABARAP by in silico docking and large-scale crystallographic fragment screening. Data based on approximately 1000 crystal structures revealed that most fragments bound to the HP2 but not to the HP1 pocket within the LIR docking site, suggesting a favorable ligandability of HP2. Through this study, we identified diverse validated LC3/GABARAP ligands and fragments as starting points for chemical probe and ATTEC development.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- 424228829/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 401883058/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 03ZU1109XX/Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)
- 03ZU1109XX/Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)
- 03ZU1109XX/Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)
LinkOut - more resources
Full Text Sources

