Haploinsufficiency at the CX3CR1 locus of hematopoietic stem cells favors the appearance of microglia-like cells in the central nervous system of transplant recipients
- PMID: 39587072
- PMCID: PMC11589136
- DOI: 10.1038/s41467-024-54515-4
Haploinsufficiency at the CX3CR1 locus of hematopoietic stem cells favors the appearance of microglia-like cells in the central nervous system of transplant recipients
Abstract
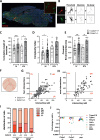
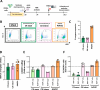
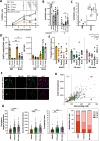
Transplantation of engineered hematopoietic stem/progenitor cells (HSPCs) showed curative potential in patients affected by neurometabolic diseases treated in early stage. Favoring the engraftment and maturation of the engineered HSPCs in the central nervous system (CNS) could allow enhancing further the therapeutic potential of this approach. Here we unveil that HSPCs haplo-insufficient at the Cx3cr1 (Cx3cr1-/+) locus are favored in central nervous system (CNS) engraftment and generation of microglia-like progeny cells (MLCs) as compared to wild type (Cx3cr1+/+) HSPCs upon transplantation in mice. Based on this evidence, we have developed a CRISPR-based targeted gene addition strategy at the human CX3CR1 locus resulting in an enhanced ability of the edited human HSPCs to generate mature MLCs upon transplantation in immunodeficient mice, and in lineage specific, regulated and robust transgene expression. This approach, which benefits from the modulation of pathways involved in microglia maturation and migration in haplo-insufficient cells, may broaden the application of HSPC gene therapy to a larger spectrum of neurometabolic and neurodegenerative diseases.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: A.B., A.M., and P.G. are co-authors in the provisional patent application PCT/US23/22029. A.B. and A.M. are co-authors in the patent PCT/US20/45106. The remaining authors declare no competing interests.
Figures



References
-
- Biffi, A. et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science341, 1233158 (2013). - PubMed
-
- Sessa, M. et al. Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: an ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet388, 476–487 (2016). - PubMed
-
- Gentner, B. et al. Hematopoietic stem- and progenitor-cell gene therapy for Hurler syndrome. N. Engl. J. Med.385, 1929–1940 (2021). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

