Characterization of the landscape of the intratumoral microbiota reveals that Streptococcus anginosus increases the risk of gastric cancer initiation and progression
- PMID: 39587089
- PMCID: PMC11589709
- DOI: 10.1038/s41421-024-00746-0
Characterization of the landscape of the intratumoral microbiota reveals that Streptococcus anginosus increases the risk of gastric cancer initiation and progression
Abstract
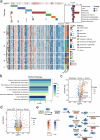
As a critical component of the tumour immune microenvironment (TIME), the resident microbiota promotes tumorigenesis across a variety of cancer types. Here, we integrated multiple types of omics data, including microbiome, transcriptome, and metabolome data, to investigate the functional role of intratumoral bacteria in gastric cancer (GC). The microbiome was used to categorize GC samples into six subtypes, and patients with a high abundance of Streptococcus or Pseudomonas had a markedly worse prognosis. Further assays revealed that Streptococcus anginosus (SA) promoted tumour cell proliferation and metastasis while suppressing the differentiation and infiltration of CD8+ T cells. However, antibiotic treatment significantly suppressed tumorigenesis in SA+ mice in vivo. We further demonstrated that the SA arginine pathway increased the abundance of ornithine, which may be a major contributor to reshaping of the TIME. Our findings demonstrated that SA, a novel risk factor, plays significant roles in the initiation and progression of GC, suggesting that SA might be a promising target for the diagnosis and treatment of GC.
© 2024. The Author(s).
Conflict of interest statement
Conflict of interest: The authors declare no competing interests.
Figures






References
-
- Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

