High-throughput genotyping of Plasmodium vivax in the Peruvian Amazon via molecular inversion probes
- PMID: 39587110
- PMCID: PMC11589703
- DOI: 10.1038/s41467-024-54731-y
High-throughput genotyping of Plasmodium vivax in the Peruvian Amazon via molecular inversion probes
Abstract
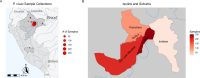
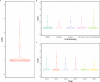
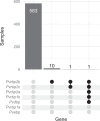
Plasmodium vivax transmission occurs throughout the tropics and is an emerging threat in areas of Plasmodium falciparum decline, causing relapse infections that complicate treatment and control. Targeted sequencing for P. falciparum has been widely deployed to detect population structure and the geographic spread of antimalarial and diagnostic resistance. However, there are fewer such tools for P. vivax. Leveraging global variation data, we designed four molecular inversion probe (MIP) genotyping panels targeting geographically differentiating SNPs, neutral SNPs, putative antimalarial resistance genes, and vaccine candidate genes. We deployed these MIP panels on 866 infections from the Peruvian Amazon and identified transmission networks with clonality (IBD[identity by descent]>0.99), copy number variation in Pvdbp and multiple Pvrbps, mutations in antimalarial resistance orthologs, and balancing selection in 13 vaccine candidate genes. Our MIP panels are the broadest genotyping panel currently available and are poised for successful deployment in other regions of P. vivax transmission.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Update of
-
High-Throughput Genotyping of Plasmodium vivax in the Peruvian Amazon via Molecular Inversion Probes.medRxiv [Preprint]. 2024 Jun 28:2024.06.27.24309599. doi: 10.1101/2024.06.27.24309599. medRxiv. 2024. Update in: Nat Commun. 2024 Nov 25;15(1):10219. doi: 10.1038/s41467-024-54731-y. PMID: 38978652 Free PMC article. Updated. Preprint.
References
-
- World Health Organization. World Malaria Report 2022 (WHO, 2022).
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- R01 TW010870/TW/FIC NIH HHS/United States
- K24 AI134990/AI/NIAID NIH HHS/United States
- R01AI165537/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI165537/AI/NIAID NIH HHS/United States
- R01TW010870/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources

