Development and validation of nomogram models for severe and fatal COVID-19
- PMID: 39587251
- PMCID: PMC11589750
- DOI: 10.1038/s41598-024-80310-8
Development and validation of nomogram models for severe and fatal COVID-19
Erratum in
-
Publisher Correction: Development and validation of nomogram models for severe and fatal COVID-19.Sci Rep. 2025 Mar 18;15(1):9365. doi: 10.1038/s41598-025-90950-z. Sci Rep. 2025. PMID: 40102222 Free PMC article. No abstract available.
Abstract
Background: The coronavirus disease 2019 (COVID-19) has exhibited escalating contagion and resistance to immunity, resulting in a surge in infections and severe cases. This study endeavors to formulate two nomogram predictive models aimed at discerning patients at heightened risk of severe and fatal outcomes upon hospital admission. The primary objective is to enhance clinical management protocols and mitigate the incidence of severe illness and mortality associated with COVID-19.
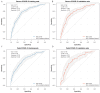
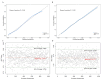
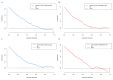
Methods: 1600 patients diagnosed with COVID-19 and discharged from Fujian Provincial Hospital were chosen as the subjects of this study. These patients were categorized into three groups: mild group (n = 940), severe group (n = 433), and fatal group (n = 227). The patients were randomly divided into training and validation cohorts in a 7:3 ratio. COVID-19 symptoms were treated as dependent variables, and univariate regression analysis was conducted for the laboratory indicators. Risk factors with p-values greater than 0.05 in the univariate regression analysis were eliminated. The remaining risk factors were then analyzed using direct multiple regression analysis to establish an unadjusted model. Subsequently, risk factors with p-values greater than 0.05 were further removed. Clinical characteristics were added to the model as adjustment factors, and the method of multiple stepwise regression analysis was employed to derive the final fully adjusted model. The severe and fatal COVID-19 models were converted into nomograms, respectively. Receiver operating characteristic (ROC) curves were utilized to evaluate the discrimination of the nomogram models. Calibration was assessed using the Hosmer-Lemeshow test and calibration curves. Clinical benefit was evaluated by decision curve analysis.
Results: Compared to the mild group, individuals in the severe COVID-19 group exhibited significant increases in age, neutrophil (NEU), and lactate dehydrogenase (LDH) levels, alongside notable decreases in lymphocyte (LYM) and albumin (ALB) levels. Nomogram model incorporating age, NEU, LDH, LYM, and ALB demonstrated efficacy in predicting the onset of severe COVID-19 (AUC = 0.771). Furthermore, history of cerebral infarction and cancer, LDH and ALB as risk factors for fatal COVID-19 cases compared to the severe group. The nomogram model comprising these factors was capable of early identification of COVID-19 fatalities (AUC = 0.748).
Conclusions: Elevated age, NEU, and LDH levels, along with decreased LYM and albumin (ALB) levels, are risk factors for severe illness in hospitalized patients with COVID-19. A history of cerebral infarction and tumors, along with elevated LDH and decreased ALB levels, are risk factors for death in critically ill patients. The nomogram model based on these factors can effectively predict the risk of severe or fatal illness from COVID-19, thereby assisting clinicians in timely interventions to reduce the rates of severe illness and mortality among hospitalized patients. However, the model faces challenges in processing longitudinal data and specific points in time, indicating that there is room for improvement.
Keywords: COVID-19; Development; Nomogram model; Risk factors; Validation.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Fujian Provincial Hospital. The data used in this study were anonymized before use. Informed consent: was obtained from all individual participants included in the study.
Figures

References
-
- Bloom, C. I. et al. ISARIC investigators. Risk of adverse outcomes in patients with underlying respiratory conditions admitted to hospital with COVID-19: a national, multicentre prospective cohort study using the ISARIC WHO Clinical Characterisation Protocol UK. Lancet Respir Med.9 (7), 699–711 (2021). - PMC - PubMed
-
- Grimes, D. A. The nomogram epidemic: resurgence of a medical relic. Ann. Intern. Med.149 (4), 273–275 (2008). - PubMed
-
- World Health Organization. Tracking SARS-CoV-2 variants. https://www.who.int/activities/tracking-SARS-CoV-2-variants
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

