Macrophages and nociceptor neurons form a sentinel unit around fenestrated capillaries to defend the synovium from circulating immune challenge
- PMID: 39587345
- PMCID: PMC11588661
- DOI: 10.1038/s41590-024-02011-8
Macrophages and nociceptor neurons form a sentinel unit around fenestrated capillaries to defend the synovium from circulating immune challenge
Abstract
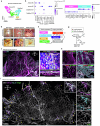
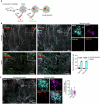
A wide variety of systemic pathologies, including infectious and autoimmune diseases, are accompanied by joint pain or inflammation, often mediated by circulating immune complexes (ICs). How such stimuli access joints and trigger inflammation is unclear. Whole-mount synovial imaging revealed PV1+ fenestrated capillaries at the periphery of the synovium in the lining-sublining interface. Circulating ICs extravasated from these PV1+ capillaries, and nociceptor neurons and three distinct macrophage subsets formed a sentinel unit around them. Macrophages showed subset-specific responses to systemic IC challenge; LYVE1+CX3CR1+ macrophages orchestrated neutrophil recruitment and activated calcitonin gene-related peptide+ (CGRP+) nociceptor neurons via interleukin-1β. In contrast, major histocompatibility complex class II+CD11c+ (MHCII+CD11c+) and MHCII+CD11c- interstitial macrophages formed tight clusters around PV1+ capillaries in response to systemic immune stimuli, a feature enhanced by nociceptor-derived CGRP. Altogether, we identify the anatomical location of synovial PV1+ capillaries and subset-specific macrophage-nociceptor cross-talk that forms a blood-joint barrier protecting the synovium from circulating immune challenges.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures















References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

