Immune landscape of the tumour microenvironment in Ethiopian breast cancer patients
- PMID: 39587630
- PMCID: PMC11587711
- DOI: 10.1186/s13058-024-01916-4
Immune landscape of the tumour microenvironment in Ethiopian breast cancer patients
Abstract
Background: The clinical management of breast cancer (BC) is mainly based on the assessment of receptor expression by tumour cells. However, there is still an unmet need for novel biomarkers important for prognosis and therapy. The tumour immune microenvironment (TIME) is thought to play a key role in prognosis and therapy selection, therefore this study aimed to describe the TIME in Ethiopian BC patients.
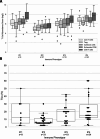
Methods: RNA was isolated from formalin-fixed paraffin-embedded (FFPE) tissue from 82 women with BC. Expression of PAM50 and 54 immune genes was analysed using the Nanostring platform and differentially expressed genes (DEGs) were determined using ROSALIND®. The abundance of different cell populations was estimated using Nanostring's cell type profiling module, while tumour infiltrating lymphocytes (TILs) were analysed using haematoxylin and eosin (H&E) staining. In addition, the PIK3CA gene was genotyped for three hotspot mutations using qPCR. Kaplan-Meier survival analysis and log-rank test were performed to compare the prognostic relevance of immune subgroups.
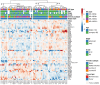
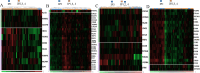
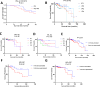
Results: Four discrete immune phenotypes (IP1-4) were identified through hierarchical clustering of immune gene expression data. These IPs were characterized by DEGs associated with both immune activation and inhibition as well as variations in the extent of immune infiltration. However, there were no significant differences regarding PIK3CA mutations between the IPs. A downregulation of immune suppressive and activating genes and the lowest number of infiltrating immune cells were found in IP2, which was associated with luminal tumours. In contrast, IP4 displayed an active TME chracterized by an upregulation of cytotoxic genes and the highest density of immune cell infiltrations, independent of the specific intrinsic subtype. IP1 and IP3 exhibited intermediate characteristics. The IPs had a prognostic relevance and patients with an active TME had improved overall survival compared to IPs with a significant downregulation of the majority of immune genes.
Conclusion: Immune gene expression profiling identified four distinct immune contextures of the TME with unique gene expression patterns and immune infiltration. The classification into distinct immune subgroups may provide important information regarding prognosis and the selection of patients undergoing conventional treatments or immunotherapies.
Keywords: Breast cancer; Ethiopia; Immune phenotypes; PAM50; Tumour immune microenvironment.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was conducted in accordance with the principles outlined in the Declaration of Helsinki. It was approved by the institutional review board of collage of health sciences of Addis Ababa University (Protocol Number DMIP 092/17/17) and National ethics review committee of Ethiopia (Protocol MOSHE/RD). In addition, sample was collected after written informed consent was obtained from participants. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. - PubMed
-
- Perou CM, Sørile T, Eisen MB, Van De Rijn M, Jeffrey SS, Ress CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. - PubMed
MeSH terms
Substances
Grants and funding
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- GTDR16378013/KOMEN/Susan G. Komen/United States
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- 2018_HA31SP/Else Kröner-Fresenius-Zentrum für Ernährungsmedizin
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 57216764/Deutscher Akademischer Austauschdienst
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
- ID 81256434/Deutsche Gesellschaft für Internationale Zusammenarbeit
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

