Mechanism of efficacy of trabectedin against myxoid liposarcoma entails detachment of the FUS-DDIT3 transcription factor from its DNA binding sites
- PMID: 39587691
- PMCID: PMC11590625
- DOI: 10.1186/s13046-024-03228-z
Mechanism of efficacy of trabectedin against myxoid liposarcoma entails detachment of the FUS-DDIT3 transcription factor from its DNA binding sites
Abstract
Background: The marine drug trabectedin has shown unusual effectiveness in the treatment of myxoid liposarcoma (MLPS), a liposarcoma characterized by the expression of the FUS-DDIT3 chimera. Trabectedin elicits a significant transcriptional response in MLPS resulting in cellular depletion and reactivation of adipogenesis. However, the role of the chimeric protein in the mechanism of action of the drug is not entirely understood.
Methods: FUS-DDIT3-specific binding sites were assessed through Chromatin Immunoprecipitation Sequencing (ChIP-Seq). Trabectedin-induced effects were studied on pre-established patient-derived xenograft models of MLPS, one sensitive to (ML017) and one resistant against (ML017ET) trabectedin at different time points (24 and 72 h, 15 days). Data were integrated with RNA-Seq from the same models.
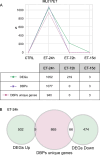
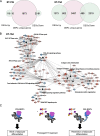
Results: Through ChIP-Seq, here we demonstrate that trabectedin inhibits the binding of FUS-DDIT3 to its target genes, restoring adipocyte differentiation in a patient-derived xenograft model of MLPS sensitive to trabectedin. In addition, complementary RNA-Seq data on the same model demonstrates a two-phase effect of trabectedin, characterized by an initial FUS-DDIT3-independent cytotoxicity, followed by a transcriptionally active pro-differentiation phase due to the long-lasting detachment of the chimera from the DNA. Interestingly, in a trabectedin-resistant MLPS model, the effect of trabectedin on FUS-DDIT3 rapidly decreased over time, and prolonged treatment was no longer able to induce any transcription or post-transcriptional modifications.
Conclusions: These findings explain the unusual mechanism underlying trabectedin's effectiveness against MLPS by pinpointing the chimera's role in inducing the differentiation block responsible for MLPS pathogenesis. Additionally, the findings hint at a potential mechanism of resistance acquired in vivo.
Keywords: Adipogenesis; Chromatin immunoprecipitation sequencing; Heterografts; Liposarcoma; Myxoid; Recombinant fusion proteins; Trabectedin.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Procedures involving animals and their care were conducted in conformity with the Italian Governing Law (D.lgs 26/2014; Authorization n.19/2008-A issued March 6, 2008, by the Ministry of Health), Mario Negri Institutional Regulations and Policies providing internal authorization for persons conducting animal experiments (Quality Management System Certificate—UNI EN ISO 9001:2008—Reg. No. 6121), the NIH Guide for the Care and Use of Laboratory Animals (2011 edition) and EU directives and guidelines (EEC Council Directive 2010/63/UE) and guidelines for the welfare and use of animals in cancer research. Consent for publication: All authors read this manuscript and approve for publication. Competing interests: The authors declare that they have no competing interests.
Figures



References
-
- D’Incalci M, Galmarini CM. A review of trabectedin (ET-743): a unique mechanism of action. Mol Cancer Ther. 2010;9:2157–63. - PubMed
-
- EMA. Yondelis. European Medicines Agency. 2018. Available from: https://www.ema.europa.eu/en/medicines/human/EPAR/yondelis. Cited 2023 Aug 23.
-
- Barone A, Chi D-C, Theoret MR, Chen H, He K, Kufrin D, et al. FDA Approval Summary: Trabectedin for Unresectable or Metastatic Liposarcoma or Leiomyosarcoma Following an Anthracycline-Containing Regimen. Clin Cancer Res. 2017;23:7448–53. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

