Comparison of Crohn's disease-associated adherent-invasive Escherichia coli (AIEC) from France and Hong Kong: results from the Pacific study
- PMID: 39587720
- PMCID: PMC11601055
- DOI: 10.1080/19490976.2024.2431645
Comparison of Crohn's disease-associated adherent-invasive Escherichia coli (AIEC) from France and Hong Kong: results from the Pacific study
Abstract
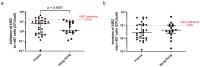
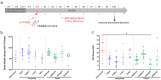
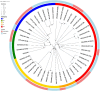
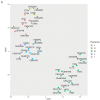
Association between ileal colonization by Adherent-Invasive Escherichia coli (AIEC) and Crohn's disease (CD) has been widely described in high-incidence Western countries but remains unexplored in Asian countries with a fast increase in CD incidence. In the PACIFIC study, we compared the characteristics of AIEC pathobionts retrieved from ileal biopsies of CD patients enrolled in France (FR) and Hong Kong (HK). The prevalence of AIEC was similar in France (24.5%, 25/102) and Hong Kong (30.0%, 18/60) (p = 0.44). No difference was observed between the two populations of AIEC regarding adhesion and invasion levels. When tested for antibiotic resistance, the proportion of AIEC strains resistant to ampicillin, piperacillin, tobramycin, and gentamicin was significantly higher in HK AIEC strains compared to French strains. AIEC strains from FR or HK population were both able to persist in the mice intestine (DSS-treated CEABAC10 mice model). Moreover, genomic analysis of 25 FR and 17 hK AIEC strains using next-generation sequencing revealed the co-existence of several virulence factors associated with enteric E. coli pathotypes, although no single virulence factor was significantly associated with either country of origin or AIEC status. In vitro, all AIEC strains (FR and HK) were sensitive to the EcoActive™ phage cocktail, suggesting that it could be a promising option to target AIEC in CD across the world.
Keywords: Crohn’s disease; adherent-invasive E. coli; bacterial comparison; trans-ethnic study.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Pariente B, Mary J-Y, Danese S, Chowers Y, De Cruz P, D’Haens G, Loftus EV, Louis E, Panés J, Schölmerich J, et al. Development of the Lemann index to assess digestive tract damage in patients with Crohn’s disease. Gastroenterology. 2015;148:52–63.e3. doi:10.1053/j.gastro.2014.09.015. - DOI - PubMed
-
- Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142(1):46–54.e42. quiz e30. doi:10.1053/j.gastro.2011.10.001. - DOI - PubMed
-
- Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st Century: a systematic review of population-based studies. Lancet Lond Engl. 2017;390(10114):2769–20. doi:10.1016/S0140-6736(17)32448-0. - DOI - PubMed
-
- Buie MJ, Quan BKin J, Windsor JW, Coward S, Hansen TM, King JA, Kotze PG, Gearry RB, Ng SC, Mak JW, et al. GLobal hospitalization trends for Crohn’s disease and ulcerative colitis: systematic review with temporal analyses. Clin Gastroenterol Hepatol. 2022;162(3):S45–S46. doi:10.1053/j.gastro.2021.12.094. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
