Sequence and Configuration of a Novel Bispecific Antibody Format Impacts Its Production Using Chinese Hamster Ovary (CHO) Cells
- PMID: 39587782
- PMCID: PMC11718431
- DOI: 10.1002/bit.28879
Sequence and Configuration of a Novel Bispecific Antibody Format Impacts Its Production Using Chinese Hamster Ovary (CHO) Cells
Abstract
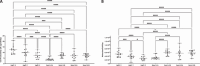
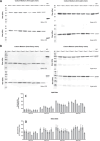
There are a number of new format antibody-inspired molecules with multiple antigen binding capabilities in development and clinical evaluation. Here, we describe the impact of the sequence and configuration of a unique bispecific antibody format (termed BYbe) using a panel of four BYbe's and the three IgG1s from which they were derived on their production in a Chinese hamster ovary (CHO) cell expression system. Following transfection and selection, one bispecific antibody format yielded fewer mini-pools in comparison to the other bispecific cell pools. When the top 12 expressing stable mini-pools of all BYbe configurations and sequences were evaluated, both the dsscFv sequence and antibody chain configuration or placement directly impacted productivity. The cell-specific productivity (qP, pg/cell/day) was lower in all BYbe cell pools compared to the IgG1 cell lines. However, when the actual molecules/cell/day produced were considered, three of the four bispecific cell pools outproduced the parental IgG1 cell pools. While gene copy number did not correlate to productivity, mRNA analysis showed that for specific BYbe formats there was a strong correlation with productivity. In summary, we describe how bispecific antibody format configuration impacts the cell line construction process and yield of product from CHO cells.
Keywords: BYbe; CHO cells; bispecific antibodies; cell line construction; recombinant antibodies.
© 2024 The Author(s). Biotechnology and Bioengineering published by Wiley Periodicals LLC.
Conflict of interest statement
Mark Ellis, Matthew Hinchliffe, David P. Humphreys, and James White are employees of UCB Pharma. Paul E. Stephens and Bernie Sweeney are former employees of UCB Pharma. The other authors declare no conflicts of interest.
Figures




References
-
- Barnes, L. M. , Bentley C. M., and Dickson A. J.. 2003. “Stability of Protein Production From Recombinant Mammalian Cells.” Biotechnology and Bioengineering 81: 631–639. - PubMed
-
- Barnes, L. M. , and Dickson A. J.. 2006. “Mammalian Cell Factories for Efficient and Stable Protein Expression.” Current Opinion in Biotechnology 17: 381–386. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

