Upregulated miR-10b-5p as a potential miRNA signature in amyotrophic lateral sclerosis patients
- PMID: 39588282
- PMCID: PMC11586771
- DOI: 10.3389/fncel.2024.1457704
Upregulated miR-10b-5p as a potential miRNA signature in amyotrophic lateral sclerosis patients
Abstract
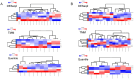
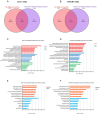
Amyotrophic lateral sclerosis (ALS) is a fatal, adult-onset disease marked by a progressive degeneration of motor neurons (MNs) present in the spinal cord, brain stem and motor cortex. Death in most patients usually occurs within 2-4 years after symptoms onset. Despite promising progress in delineating underlying mechanisms, such as disturbed proteostasis, DNA/RNA metabolism, splicing or proper nucleocytoplasmic shuttling, there are no effective therapies for the vast majority of cases. A reason for this might be the disease heterogeneity and lack of substantial clinical and molecular biomarkers. The identification and validation of such pathophysiology driven biomarkers could be useful for early diagnosis and treatment stratification. Recent advances in next generation RNA-sequencing approaches have provided important insights to identify key changes of non-coding RNAs (ncRNAs) implicated with ALS disease. Especially, microRNAs (miRNAs) have emerged as key post-transcriptional regulators of gene expression to target several genes/pathways by degrading messenger RNAs (mRNAs) or repressing levels of gene expression. In this study, we expand our previous work to identify top-regulated differentially expressed (DE)-miRNAs by combining different normalizations to search for important and generalisable pathomechanistic dysregulations in ALS as putative novel biomarkers of the disease. For this we performed a consensus pipeline of existing datasets to investigate the transcriptomic profile (mRNAs and miRNAs) of MN cell lines from iPSC-derived SOD1- and TARDBP (TDP-43 protein)-mutant-ALS patients and healthy controls to identify potential signatures and their related pathways associated with neurodegeneration. Transcriptional profiling of miRNA-mRNA interactions from MN cell lines in ALS patients revealed differential expression of genes showed greater vulnerability to KEAP1-NRF2 stress response pathway, sharing a common molecular denominator linked to both disease conditions. We also reported that mutations in above genes led to significant upregulation of the top candidate miR-10b-5p, which we could validate in immortalized lymphoblast cell lines (LCLs) derived from sporadic and familial ALS patients and postmortem tissues of familial ALS patients. Collectively, our findings suggest that miRNA analysis simultaneously performed in various human biological samples may reveal shared miRNA profiles potentially useful as a biomarker of the disease.
Keywords: amyotrophic lateral sclerosis; differentially expressed; human induced pluripotent stem cells; microRNA; motor neurons; next generation RNA sequencing.
Copyright © 2024 Dash, Freischmidt, Helferich, Ludolph, Andersen, Weishaupt and Hermann.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

