Shared differential factors underlying individual spontaneous neural activity abnormalities in major depressive disorder
- PMID: 39588672
- PMCID: PMC11650188
- DOI: 10.1017/S0033291724002617
Shared differential factors underlying individual spontaneous neural activity abnormalities in major depressive disorder
Abstract
Background: In contemporary neuroimaging studies, it has been observed that patients with major depressive disorder (MDD) exhibit aberrant spontaneous neural activity, commonly quantified through the amplitude of low-frequency fluctuations (ALFF). However, the substantial individual heterogeneity among patients poses a challenge to reaching a unified conclusion.
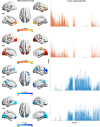
Methods: To address this variability, our study adopts a novel framework to parse individualized ALFF abnormalities. We hypothesize that individualized ALFF abnormalities can be portrayed as a unique linear combination of shared differential factors. Our study involved two large multi-center datasets, comprising 2424 patients with MDD and 2183 healthy controls. In patients, individualized ALFF abnormalities were derived through normative modeling and further deconstructed into differential factors using non-negative matrix factorization.
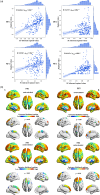
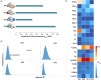
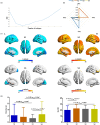
Results: Two positive and two negative factors were identified. These factors were closely linked to clinical characteristics and explained group-level ALFF abnormalities in the two datasets. Moreover, these factors exhibited distinct associations with the distribution of neurotransmitter receptors/transporters, transcriptional profiles of inflammation-related genes, and connectome-informed epicenters, underscoring their neurobiological relevance. Additionally, factor compositions facilitated the identification of four distinct depressive subtypes, each characterized by unique abnormal ALFF patterns and clinical features. Importantly, these findings were successfully replicated in another dataset with different acquisition equipment, protocols, preprocessing strategies, and medication statuses, validating their robustness and generalizability.
Conclusions: This research identifies shared differential factors underlying individual spontaneous neural activity abnormalities in MDD and contributes novel insights into the heterogeneity of spontaneous neural activity abnormalities in MDD.
Keywords: amplitude of low-frequency fluctuations; dimension; heterogeneity; major depressive disorder; normative modeling.
Conflict of interest statement
None.
Figures




References
-
- Aghourian, M., Legault-Denis, C., Soucy, J. P., & Rosa-Neto, P. (2017). Quantification of brain cholinergic denervation in Alzheimer's disease using PET imaging with [(18)F]-FEOBV. Molecular Psychiatry, 22, 1531–1538. - PubMed
-
- Ashburner, J. (2009). Computational anatomy with the SPM software. Magnetic Resonance Imaging, 27, 1163–1174. - PubMed
-
- Azen, R., & Budescu, D. V. (2003). The dominance analysis approach for comparing predictors in multiple regression. Psychological Methods, 8, 129–148. - PubMed
-
- Baldassarri, S. R., Hillmer, A. T., Anderson, J. M., Jatlow, P., Nabulsi, N., Labaree, D., … Esterlis, I. (2018). Use of electronic cigarettes leads to significant beta2-nicotinic acetylcholine receptor occupancy: Evidence from a PET imaging study. Nicotine & Tobacco Research, 20, 425–433. - PMC - PubMed
LinkOut - more resources
Full Text Sources

