Retargeting target-directed microRNA-decay sites to highly expressed viral or cellular miRNAs
- PMID: 39588775
- PMCID: PMC11662681
- DOI: 10.1093/nar/gkae1103
Retargeting target-directed microRNA-decay sites to highly expressed viral or cellular miRNAs
Abstract
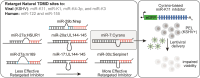
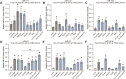
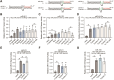
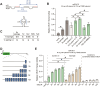
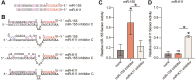
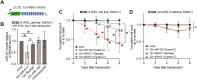
MicroRNAs (miRNAs) are pervasive regulators of gene expression, necessitating the development of tools to inhibit individual miRNAs for functional studies or therapeutic targeting. Specialized base-pairing configurations between a miRNA and an RNA target site can trigger the degradation of the targeting miRNA through target-directed miRNA decay (TDMD). Previous work has identified several natural sites that induce TDMD of specific miRNAs. We explored retargeting known TDMD sites for the inhibition of heterologous miRNAs, including several encoded by Kaposi's Sarcoma-associated herpesvirus (KSHV). We focused particularly on miR-K11, a viral mimic of the oncogenic miRNA miR-155. miRNA pairing architectures based on the TDMD site in the long non-coding RNA Cyrano outperformed other retargeted sites. Cyrano-like inhibitors were specific for viral miR-K11 over cellular miR-155 and vice versa. Lentiviral delivery of a Cyrano-like miR-K11 inhibitor into KSHV-transformed primary effusion lymphoma (PEL) cells impaired their viability, showing that miR-K11 promotes KSHV-dependent PEL cell survival. Surprisingly, inactivation of ZSWIM8, a key mediator of TDMD, did not substantially affect miRNA inhibition by retargeted Cyrano-based inhibitors in 293T or PEL cells. Together, our results demonstrate the feasibility of retargeting natural TDMD sites to highly expressed viral or cellular miRNAs and further define features of effective encoded miRNA inhibitors.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

