Chemotherapeutic agents and leucine deprivation induce codon-biased aberrant protein production in cancer
- PMID: 39588782
- PMCID: PMC11662694
- DOI: 10.1093/nar/gkae1110
Chemotherapeutic agents and leucine deprivation induce codon-biased aberrant protein production in cancer
Abstract
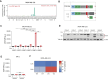
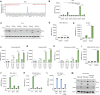
Messenger RNA (mRNA) translation is a tightly controlled process frequently deregulated in cancer. Key to this deregulation are transfer RNAs (tRNAs), whose expression, processing and post-transcriptional modifications are often altered in cancer to support cellular transformation. In conditions of limiting levels of amino acids, this deregulated control of protein synthesis leads to aberrant protein production in the form of ribosomal frameshifting or misincorporation of non-cognate amino acids. Here, we studied leucine, an essential amino acid coded by six different codons. Surprisingly, we found that leucine deprivation leads to ribosomal stalling and aberrant protein production in various cancer cell types, predominantly at one codon, UUA. Similar effects were observed after treatment with chemotherapeutic agents, implying a shared mechanism controlling the downstream effects on mRNA translation. In both conditions, a limitation in the availability of tRNALeu(UAA) for protein production was shown to be the cause for this dominant effect on UUA codons. The induced aberrant proteins can be processed and immune-presented as neoepitopes and can direct T-cell killing. Altogether, we uncovered a novel mode of interplay between DNA damage, regulation of tRNA availability for mRNA translation and aberrant protein production in cancer that could be exploited for anti-cancer therapy.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Kochavi A., Lovecchio D., Faller W.J., Agami R.. Proteome diversification by mRNA translation in cancer. Mol. Cell. 2023; 83:469–480. - PubMed
-
- van Riggelen J., Yetil A., Felsher D.W.. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 2010; 10:301–309. - PubMed
-
- de Las Heras-Rubio A., Perucho L., Paciucci R., Vilardell J., ME L.L.. Ribosomal proteins as novel players in tumorigenesis. Cancer Metastasis Rev. 2014; 33:115–141. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

