Insights into antibiotic resistance promoted by quinolone exposure
- PMID: 39589140
- PMCID: PMC11784200
- DOI: 10.1128/aac.00997-24
Insights into antibiotic resistance promoted by quinolone exposure
Abstract
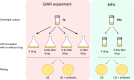
Quinolone-induced antibiotic resistance (QIAR) refers to the phenomenon by which bacteria exposed to sublethal levels of quinolones acquire resistance to non-quinolone antibiotics. We have explored this in Escherichia coli MG1655 using a variety of compounds and bacteria carrying a quinolone-resistance mutation in gyrase, mutations affecting the SOS response, and mutations in error-prone polymerases. The nature of the antibiotic-resistance mutations was determined by whole-genome sequencing. Exposure to low levels of most quinolones tested led to mutations conferring resistance to chloramphenicol, ampicillin, kanamycin, and tetracycline. The mutations included point mutations and deletions and could mostly be correlated with the resistance phenotype. QIAR depended upon DNA gyrase and involved the SOS response but was not dependent on error-prone polymerases. Only moxifloxacin, among the quinolones tested, did not display a significant QIAR effect. We speculate that the lack of QIAR with moxifloxacin may be attributable to it acting via a different mechanism. In addition to the concerns about antimicrobial resistance to quinolones and other compounds, QIAR presents an additional challenge in relation to the usage of quinolone antibacterials.
Keywords: antibiotic resistance; fluoroquinolones; gyrase; topoisomerase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- O’Neill J. 2016. Tackling drug-resistant infections globally: final report and recommendations. Available from: http://amr-review.org/Publications
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

