Permanent draft genome sequences of cadmium-resistant isolates of Cupriavidus from soils within the Tar Creek Superfund site
- PMID: 39589146
- PMCID: PMC11737155
- DOI: 10.1128/mra.00818-24
Permanent draft genome sequences of cadmium-resistant isolates of Cupriavidus from soils within the Tar Creek Superfund site
Abstract
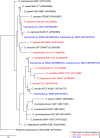
Soil samples taken near the abandoned town of Picher, OK, USA, were used to enrich and isolate bacteria in the presence of cadmium. Isolates reported belong to the genus Cupriavidus. Here, we report their permanent draft sequences with an emphasis on genes conferring resistance to cadmium.
Keywords: Cupriavidus; Tar Creek Superfund site; cadmium; cobalt; czcCBA; lead; zinc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Beattie RE, Henke W, Campa MF, Hazen TC, McAliley LR, Campbell JH. 2018. Variation in microbial community structure correlates with heavy-metal contamination in soils decades after mining ceased. Soil Biol Biochem 126:57–63. doi:10.1016/j.soilbio.2018.08.011 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
