Influenza virus infection and aerosol shedding kinetics in a controlled human infection model
- PMID: 39589151
- PMCID: PMC11657674
- DOI: 10.1128/jvi.01612-24
Influenza virus infection and aerosol shedding kinetics in a controlled human infection model
Erratum in
-
Correction for Shetty et al., "Influenza virus infection and aerosol shedding kinetics in a controlled human infection model".J Virol. 2025 Nov 25;99(11):e0135425. doi: 10.1128/jvi.01354-25. Epub 2025 Oct 7. J Virol. 2025. PMID: 41055343 Free PMC article. No abstract available.
Abstract
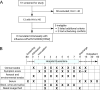
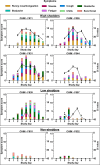
Establishing effective mitigation strategies to reduce the spread of influenza virus requires an improved understanding of the mechanisms of transmission. We evaluated the use of a controlled human infection model using an H3N2 seasonal influenza virus to study critical aspects of transmission, including symptom progression and the dynamics of virus shedding. Eight volunteers were challenged with influenza A/Perth/16/2009 (H3N2) virus between July and September 2022 at Emory University Hospital. Viral shedding in the nasopharynx, saliva, stool, urine, and respiratory aerosols was monitored over the quarantine period, and symptoms were tracked until day 15. In addition, environmental swabs were collected from participant rooms to examine fomite contamination, and participant sera were collected to assess seroconversion by hemagglutination inhibition or microneutralization assays. Among the eight participants, influenza virus infection was confirmed in six (75%). Infectious virus or viral RNA was found in multiple physiological compartments, fecal samples, aerosol particles, and on surfaces in the immediate environment. Illness was moderate, with upper respiratory symptoms dominating. In participants with the highest viral loads, antibody titers rose by day 15 post-inoculation, while in participants with low or undetectable viral loads, there was little or no increase in functional antibody titers. These data demonstrate the safety and utility of the human infection model to study features critical to influenza virus transmission dynamics in a controlled manner and will inform the design of future challenge studies focused on modeling and limiting transmission.CLINICAL TRIALSThis study is registered with ClinicalTrials.gov as NCT05332899.
Importance: We use a controlled human infection model to assess respiratory and aerosol shedding kinetics to expand our knowledge of influenza infection dynamics and help inform future studies aimed at understanding human-to-human transmission.
Keywords: aerosols; antibody response; controlled human infection model; influenza; influenza-like illness; virus shedding.
Conflict of interest statement
N.G.R. receives consulting fees from Krog and participates on the advisory boards of Moderna, Sanofi, Seqirus, and Pfizer. M.F.E. recieves consulting fees from Medtronic and Boston Scientific. L.C.M. receives consulting fees from MITRE.
Figures




References
-
- Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, Davey RT Jr, Taubenberger JK. 2016. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. MBio 7:e00417–16. doi: 10.1128/mBio.00417-16 - DOI - PMC - PubMed
-
- Memoli MJ, Czajkowski L, Reed S, Athota R, Bristol T, Proudfoot K, Fargis S, Stein M, Dunfee RL, Shaw PA, Davey RT, Taubenberger JK. 2015. Validation of the wild-type influenza A human challenge model H1N1pdMIST: an A(H1N1)pdm09 dose-finding investigational new drug study. Clin Infect Dis 60:693–702. doi: 10.1093/cid/ciu924 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

