A Potent, Selective, Small-Molecule Inhibitor of DHX9 Abrogates Proliferation of Microsatellite Instable Cancers with Deficient Mismatch Repair
- PMID: 39589774
- PMCID: PMC11831107
- DOI: 10.1158/0008-5472.CAN-24-0397
A Potent, Selective, Small-Molecule Inhibitor of DHX9 Abrogates Proliferation of Microsatellite Instable Cancers with Deficient Mismatch Repair
Abstract
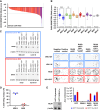
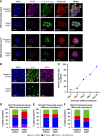
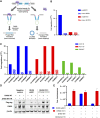
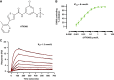
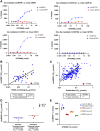
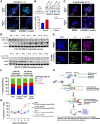
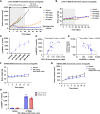
DHX9 is a multifunctional DExH-box RNA helicase with important roles in the regulation of transcription, translation, and maintenance of genome stability. Elevated expression of DHX9 is evident in multiple cancer types, including colorectal cancer. Microsatellite instable-high (MSI-H) tumors with deficient mismatch repair (dMMR) display a strong dependence on DHX9, making this helicase an attractive target for oncology drug discovery. In this report, we show that DHX9 knockdown increased RNA/DNA secondary structures and replication stress, resulting in cell-cycle arrest and the onset of apoptosis in cancer cells with MSI-H/dMMR. ATX968 was identified as a potent and selective inhibitor of DHX9 helicase activity. Chemical inhibition of DHX9 enzymatic activity elicited similar selective effects on cell proliferation as seen with genetic knockdown. In addition, ATX968 induced robust and durable responses in an MSI-H/dMMR xenograft model but not in a microsatellite stable/proficient MMR model. These preclinical data validate DHX9 as a target for the treatment of patients with MSI-H/dMMR. Additionally, this potent and selective inhibitor of DHX9 provides a valuable tool with which to further explore the effects of inhibition of DHX9 enzymatic activity on the proliferation of cancer cells in vitro and in vivo. Significance: DHX9 is required in cancer cells with deficient mismatch repair and can be inhibited by ATX968, providing a promising strategy for the development of precision cancer therapeutics.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
J. Castro reports patents for WO/2023/154519 and W0/2024/220373 issued. M.H. Daniels reports a patent for WO/2023/154519 pending. D. Brennan reports a patent for W0/2024/220373 pending. Y.-T. Lee reports a patent for WO/2023/154519 issued. B.A. Sparling reports a patent for WO/2023/154519 issued. E.A. Sickmier reports a patent for WO/2023/154519 issued. S.J. Blakemore reports other support from Accent Therapeutics during the conduct of the study. P.A. Boriack-Sjodin reports being an employee and stockholder of Accent Therapeutics. K.W. Duncan reports a patent for WO/2023/154519 pending. S. Ribich reports patents for WO/2023/154519 and W0/2024/220373 issued and pending, respectively. R.A. Copeland reports personal fees from Atlas Venture, Rectify Pharma, and Triana Biomedicines outside the submitted work and is an employee and shareholder of Accent Therapeutics, Inc. No disclosures were reported by the other authors.
Figures

References
-
- Wells JP, White J, Stirling PC. R loops and their composite cancer connections. Trends Cancer 2019;5:619–31. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

