Role of cGAS/STING pathway in aging and sexual dimorphism in diabetic kidney disease
- PMID: 39589791
- PMCID: PMC11721291
- DOI: 10.1172/jci.insight.174126
Role of cGAS/STING pathway in aging and sexual dimorphism in diabetic kidney disease
Abstract
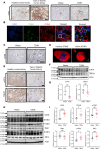
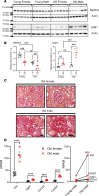
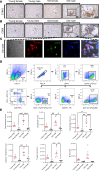
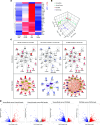
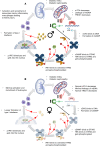
Diabetic kidney disease (DKD) is the leading cause of chronic renal pathology. Understanding the molecular underpinnings of DKD is critical to designing tailored therapeutic approaches. Here, we focused on sex differences and the contribution of aging toward the progression of DKD. To explore these questions, we utilized young (12 weeks old) and aged (approximately 50 weeks old) type 2 diabetic nephropathy (T2DN) rats. We revealed that the cyclic GMP-AMP synthase (cGAS)/stimulator of interferon genes (STING) pathway was upregulated in T2DN rats compared with nondiabetic Wistar rats and in type 2 diabetic human kidneys. The activation of the cGAS/STING signaling pathway exhibited distinct protein expression profiles between male and female T2DN rats, with these differences becoming more pronounced with aging. RNA-Seq analysis of the kidney cortex in both male and female T2DN rats, at both younger and older ages, revealed several key molecules, highlighting crucial genes within the cGAS/STING pathway. Thus, our study delved deep into understanding the intricate sexual differences in the development and progression of DKD and we propose the cGAS/STING pathway as an essential contributor to disease development.
Keywords: Chronic kidney disease; Diabetes; Nephrology.
Conflict of interest statement
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

