Endogenous antigens shape the transcriptome and TCR repertoire in an autoimmune arthritis model
- PMID: 39589811
- PMCID: PMC11735108
- DOI: 10.1172/JCI174647
Endogenous antigens shape the transcriptome and TCR repertoire in an autoimmune arthritis model
Abstract
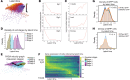
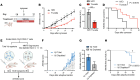
The development of pathogenic autoreactive CD4+ T cells, particularly in the context of impaired signaling, remains poorly understood. Unraveling how defective signaling pathways contribute to their activation and persistence is crucial for identifying new therapeutic targets. We performed bulk and single-cell RNA-Seq (scRNA-Seq) and single-cell T cell receptor sequencing (scTCR-Seq) to profile a highly arthritogenic subset of naive CD4+ T cells from BALB/c-Zap70*W163C (SKG) mice, which develop CD4+ T cell-mediated autoimmune arthritis driven by a hypomorphic mutation in Zap70 - a key TCR signaling kinase. Despite impaired signaling, these cells exhibited heightened expression of T cell activation and cytokine signaling genes but diminished expression of a subset of tolerogenic markers (Izumo1r, Tnfrsf9, Cd5, S100a11) compared with WT cells. The arthritogenic cells showed an enrichment for TCR variable β (Vβ) chains targeting superantigens (Sags) from the endogenous mouse mammary tumor virus (MMTV) but exhibited diminished induction of tolerogenic markers following peripheral antigen encounter, contrasting with the robust induction of the negative regulators seen in WT cells. In arthritic joints, cells expressing Sag-reactive Vβs expanded alongside detectable MMTV proviruses. Antiretroviral treatment and Sag-reactive T cell depletion curtailed SKG arthritis, suggesting that endogenous retroviruses disrupted peripheral tolerance and promoted the activation and differentiation of autoreactive CD4+ T cells into pathogenic effector cells.
Keywords: Autoimmune diseases; Autoimmunity; Immunology; T cell receptor; Tolerance.
Conflict of interest statement
Figures







Comment in
- A new twist on superantigen-activated autoimmune disease doi: 10.1172/JCI187567
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

