MR1 presents vitamin B6-related compounds for recognition by MR1-reactive T cells
- PMID: 39589872
- PMCID: PMC11626183
- DOI: 10.1073/pnas.2414792121
MR1 presents vitamin B6-related compounds for recognition by MR1-reactive T cells
Abstract
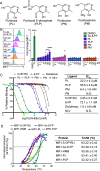
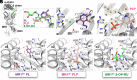
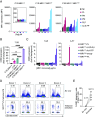
The major histocompatibility complex class I related protein (MR1) presents microbially derived vitamin B2 precursors to mucosal-associated invariant T (MAIT) cells. MR1 can also present other metabolites to activate MR1-restricted T cells expressing more diverse T cell receptors (TCRs), some with anti-tumor reactivity. However, knowledge of the range of the antigen(s) that can activate diverse MR1-reactive T cells remains incomplete. Here, we identify pyridoxal (vitamin B6) as a naturally presented MR1 ligand using unbiased mass spectrometry analyses of MR1-bound metabolites. Pyridoxal, and the related compound, pyridoxal 5-phosphate bound to MR1 and enabled cell surface upregulation of wild type MR1*01 and MR1 expressing the Arg9His polymorphism associated with the MR1*04 allotype in a manner dependent on Lys43-mediated Schiff-base formation. Crystal structures of MR1*01 in complex with pyridoxal and pyridoxal 5-phosphate showed how these ligands were accommodated within the A-pocket of MR1. T cell lines transduced with the 7.G5 TCR, which has reported "pan-cancer" specificity, were specifically activated by pyridoxal presented by antigen-presenting cells expressing MR1*01 and MR1 allotypes bearing the less common Arg9His polymorphism. 7.G5 T cells also recognized, to a lesser extent, pyridoxal 5-phosphate and, importantly, recognition of both vitamers was blocked by an anti-MR1 antibody. 7.G5 TCR reactivity toward pyridoxal was enhanced when presented by the Arg9His polymorphism-bearing MR1 allotypes. Vitamin B6, and vitamers thereof, have been associated with various cancers, and here we describe a link between this ligand, MR1, and its allomorphs, and the pan-cancer 7.G5 TCR. This work identifies an MR1 ligand that can activate a diverse MR1-restricted TCR.
Keywords: MHC class I-related molecule (MR1); T cell receptor; mass spectrometry; metabolite; structural biology.
Conflict of interest statement
Competing interests statement:J.R., D.P.F., and J.M. are inventors on patent applications (PCT/AU2013/000742, WO2014005194; PCT/AU2015/050148, WO2015149130) describing MR1 ligands and MR1-tetramer reagents.
Figures


References
-
- Adams E. J., Luoma A. M., The adaptable major histocompatibility complex (MHC) fold: Structure and function of nonclassical and MHC class I-like molecules. Annu. Rev. Immunol. 31, 529–561 (2013). - PubMed
-
- Illing P. T., Ramarathinam S. H., Purcell A. W., New insights and approaches for analyses of immunopeptidomes. Curr. Opin. Immunol. 77, 102216 (2022). - PubMed
MeSH terms
Substances
Grants and funding
- DP220102401/Department of Education and Training | Australian Research Council (ARC)
- DE220101491/Department of Education and Training | Australian Research Council (ARC)
- DP240102905/Department of Education and Training | Australian Research Council (ARC)
- 2008981/DHAC | National Health and Medical Research Council (NHMRC)
- CE200100012/Department of Education and Training | Australian Research Council (ARC)
- 2016596/DHAC | National Health and Medical Research Council (NHMRC)
- AI148407-01A1/HHS | National Institutes of Health (NIH)
- 2009551/DHAC | National Health and Medical Research Council (NHMRC)
- 2008616/DHAC | National Health and Medical Research Council (NHMRC)
- R01 AI148407/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

