ER-tethered stress sensor CREBH regulates mitochondrial unfolded protein response to maintain energy homeostasis
- PMID: 39589874
- PMCID: PMC11626163
- DOI: 10.1073/pnas.2410486121
ER-tethered stress sensor CREBH regulates mitochondrial unfolded protein response to maintain energy homeostasis
Abstract
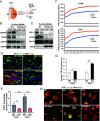
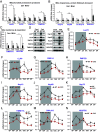
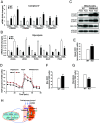
The Mitochondrial Unfolded Protein Response (UPRmt), a mitochondria-originated stress response to altered mitochondrial proteostasis, plays important roles in various pathophysiological processes. In this study, we revealed that the endoplasmic reticulum (ER)-tethered stress sensor CREBH regulates UPRmt to maintain mitochondrial homeostasis and function in the liver. CREBH is enriched in and required for hepatic Mitochondria-Associated Membrane (MAM) expansion induced by energy demands. Under a fasting challenge or during the circadian cycle, CREBH is activated to promote expression of the genes encoding the key enzymes, chaperones, and regulators of UPRmt in the liver. Activated CREBH, cooperating with peroxisome proliferator-activated receptor α (PPARα), activates expression of Activating Transcription Factor (ATF) 5 and ATF4, two major UPRmt transcriptional regulators, independent of the ER-originated UPR (UPRER) pathways. Hepatic CREBH deficiency leads to accumulation of mitochondrial unfolded proteins, decreased mitochondrial membrane potential, and elevated cellular redox state. Dysregulation of mitochondrial function caused by CREBH deficiency coincides with increased hepatic mitochondrial oxidative phosphorylation (OXPHOS) but decreased glycolysis. CREBH knockout mice display defects in fatty acid oxidation and increased reliance on carbohydrate oxidation for energy production. In summary, our studies uncover that hepatic UPRmt is activated through CREBH under physiological challenges, highlighting a molecular link between ER and mitochondria in maintaining mitochondrial proteostasis and energy homeostasis under stress conditions.
Keywords: ER-mitochondria contact; cell metabolism; michondrial UPR; transcriptional regulation; unfolded protein response.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




References
-
- Zhang K., et al. , Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124, 587–599 (2006). - PubMed
MeSH terms
Substances
Grants and funding
- R21 HL148089/HL/NHLBI NIH HHS/United States
- R01 DK132065/DK/NIDDK NIH HHS/United States
- DK110314/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- DK134361/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- R01 DK090313/DK/NIDDK NIH HHS/United States
- DK090313/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- R01 HL150474/HL/NHLBI NIH HHS/United States
- R01 DK110314/DK/NIDDK NIH HHS/United States
- DK126908/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- R01 DK134361/DK/NIDDK NIH HHS/United States
- DK132065/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- P30 DK020572/DK/NIDDK NIH HHS/United States
- R01 DK126908/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

