A Novel Machine Learning Model and a Web Portal for Predicting the Human Skin Sensitization Effects of Chemical Agents
- PMID: 39590983
- PMCID: PMC11598222
- DOI: 10.3390/toxics12110803
A Novel Machine Learning Model and a Web Portal for Predicting the Human Skin Sensitization Effects of Chemical Agents
Abstract
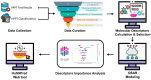
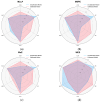
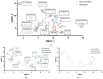
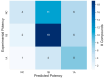
Skin sensitization is a significant concern for chemical safety assessments. Traditional animal assays often fail to predict human responses accurately, and ethical constraints limit the collection of human data, necessitating a need for reliable in silico models of skin sensitization prediction. This study introduces HuSSPred, an in silico tool based on the Human Predictive Patch Test (HPPT). HuSSPred aims to enhance the reliability of predicting human skin sensitization effects for chemical agents to support their regulatory assessment. We have curated an extensive HPPT database and performed chemical space analysis and grouping. Binary and multiclass QSAR models were developed with Bayesian hyperparameter optimization. Model performance was evaluated via five-fold cross-validation. We performed model validation with reference data from the Defined Approaches for Skin Sensitization (DASS) app. HuSSPred models demonstrated strong predictive performance with CCR ranging from 55 to 88%, sensitivity between 48 and 89%, and specificity between 37 and 92%. The positive predictive value (PPV) ranged from 84 to 97%, versus negative predictive value (NPV) from 22 to 65%, and coverage was between 75 and 93%. Our models exhibited comparable or improved performance compared to existing tools, and the external validation showed the high accuracy and sensitivity of the developed models. HuSSPred provides a reliable, open-access, and ethical alternative to traditional testing for skin sensitization. Its high accuracy and reasonable coverage make it a valuable resource for regulatory assessments, aligning with the 3Rs principles. The publicly accessible HuSSPred web tool offers a user-friendly interface for predicting skin sensitization based on chemical structure.
Keywords: NAMs; QSAR; cheminformatics; computational toxicology; skin sensitization.
Conflict of interest statement
A.T. and E.N.M. are co-founders of Predictive, LLC., which develops novel alternative methodologies and software for toxicity prediction. All the other authors declare no conflicts.
Figures




References
-
- Murphy K., Weaver C. Janeway’s Immunobiology. Garland Science; New York, NY, USA: 2017. p. 840.
-
- International Regulatory Requirements for Skin Sensitization Testing—ScienceDirect. [(accessed on 11 September 2024)]. Available online: https://www.sciencedirect.com/science/article/pii/S0273230018300667?casa....
-
- OECD . Test No. 429: Skin Sensitisation: Local Lymph Node Assay. Organisation for Economic Co-Operation and Development; Paris, France: 2010.
-
- OECD . Test No. 406: Skin Sensitisation. Organisation for Economic Co-Operation and Development; Paris, France: 2022.
-
- OECD . Guideline No. 497: Defined Approaches on Skin Sensitisation. Organisation for Economic Co-Operation and Development; Paris, France: 2023.
Grants and funding
LinkOut - more resources
Full Text Sources

