Beyond HRD Status: Unraveling Genetic Variants Impacting PARP Inhibitor Sensitivity in Advanced Ovarian Cancer
- PMID: 39591206
- PMCID: PMC11670052
- DOI: 10.1158/2767-9764.CRC-24-0294
Beyond HRD Status: Unraveling Genetic Variants Impacting PARP Inhibitor Sensitivity in Advanced Ovarian Cancer
Abstract
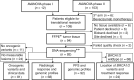
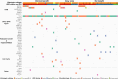
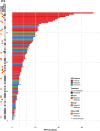
Abstract: The management of advanced epithelial ovarian cancer (AOC) has undergone significant advancements with the emergence of molecular diagnostics, particularly in predicting responses to PARP inhibitors (PARPi) based on homologous recombination deficiency (HRD) status. However, understanding sensitivity and resistance beyond HRD status remains elusive. This study aims to explore molecular factors that may elucidate why HRD status does not consistently predict PARPi sensitivity. Therefore, we conducted a post hoc translational analysis of formalin-fixed paraffin-embedded tumor samples from the ENGOT-ov24/NSGO-AVANOVA part 1 and 2 trial (NCT02354131), focusing on alterations pertaining radiologic response and progression-free survival (PFS). DNA sequencing was performed using the TruSight Oncology 500 HT gene panel, with variants classified according to recent guidelines. HRD status had been assessed by Myriad MyChoice CDx. We identified, among 92 patients in the ENGOT-ov24/NSGO-AVANOVA part 1 and 2 trial, 151 pathogenic or likely pathogenic variants across 81 samples. PARPi-sensitizing variants were found in two out of 10 HRD-negative samples from patients with clinical benefit (PFS ≥12 months), whereas three out of 10 HRD-positive samples from patients having no benefit (PFS ≤6 months) harbored variants associated with PARPi resistance. Additionally, analysis of BRCA1 variants revealed that truncating variants in exon 11 correlated with clinical benefit when niraparib was combined with bevacizumab. Conclusively, our findings highlight the complexity of PARPi response in AOC and underscore the importance of exploring somatic variants beyond HRD status. Further investigation into exon 11 variants of BRCA1 and the potential of combination treatment is warranted.
Significance: The irregular response to PARPi in HRD-positive and -negative tumors highlights the need for identifying additional biomarkers. This study explores the mutational landscape beyond HRD status in AOC, ultimately advancing precision oncology in future clinical practice.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
M.K. Kjeldsen reports grants from GSK during the conduct of the study, as well as grants from OvaCure and Knud Højgaards Fond outside the submitted work. C.A. Haslund reports personal fees from GSK, Bristol Myers Squibb, and AstraZeneca outside the submitted work. J. Mäenpää reports personal fees from Eisai outside the submitted work. T.L. Werner reports other from Repare, Blueprint, Clovis, Tesaro, and Mersana outside the submitted work. S. Hietanen reports personal fees from AstraZeneca, GSK, and Eisai outside the submitted work. H. Dahlstrand reports personal fees from Educational Sessions for AstraZeneca, Roche, and GSK outside the submitted work. L. Bjørge reports grants from AstraZeneca and other from GSK, MSD, and GSK outside the submitted work. M.R. Mirza reports grants from GSK during the conduct of the study. M. Rossing reports grants from AstraZeneca and personal fees from AstraZeneca and MSD outside the submitted work. No disclosures were reported by the other authors.
Figures


References
-
- Loverix L, Vergote I, Busschaert P, Vanderstichele A, Venken T, Boeckx B, et al. . PARP inhibitor predictive value of the Leuven HRD test compared with Myriad MyChoice CDx PLUS HRD on 468 ovarian cancer patients from the PAOLA-1/ENGOT-ov25 trial. Eur J Cancer 2023;188:131–9. - PubMed
-
- Willing EM, Vollbrecht C, Vössing C, Weist P, Schallenberg S, Herbst JM, et al. . Development of the NOGGO GIS v1 assay, a comprehensive hybrid-capture-based NGS assay for therapeutic stratification of homologous repair deficiency driven tumors and clinical validation. Cancers (Basel) 2023;15:3445. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

