Pan-cancer analysis identifies the oncogenic role of CCNE1 in human cancers
- PMID: 39591374
- PMCID: PMC11719100
- DOI: 10.18632/aging.206163
Pan-cancer analysis identifies the oncogenic role of CCNE1 in human cancers
Abstract
Objective: To investigate expression, prognosis, immune cell infiltration of Cyclin E1 (CCNE1) in cancer.
Methods: We used TIMER and GEPIA datasets to analyze the differential expression of CCNE1 in multiple tumors. GEPIA and Kaplan-Meier plotter databases were utilized to observe the prognostic significance of CCNE1 in cancer. TIMER and cBioPortal databases were adopted for the analysis regarding immune infiltration and mutation respectively.
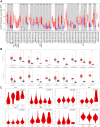
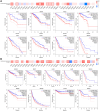
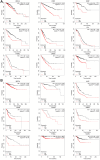
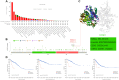
Results: The results showed that CCNE1 was highly expressed in multiple cancers including BLCA, BRCA, CHOL, COAD, ESCA, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, READ, STAD, THCA, UCEC (P < 0.001) and CESC (P < 0.01). High CCNE1 expression was associated with a poor overall survival prognosis in several cancers, including ACC, BRCA, KIRC, KIRP, LGG, LIHC, LUAD and MESO. Additionally, CCNE1 expression was correlated with the cancer-associated immune infiltration level in BRCA, COAD, LUSC, STAD and THYM.
Conclusions: CCNE1 is expected to be a potential biomarker for tumor prognosis and immune infiltration in various cancers.
Keywords: CCNE1; immune infiltration; pan-cancer; prognosis.
Conflict of interest statement
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

