Coordination between the eIF2 kinase GCN2 and p53 signaling supports purine metabolism and the progression of prostate cancer
- PMID: 39591412
- PMCID: PMC11826925
- DOI: 10.1126/scisignal.adp1375
Coordination between the eIF2 kinase GCN2 and p53 signaling supports purine metabolism and the progression of prostate cancer
Abstract
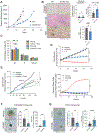
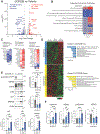
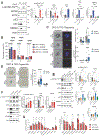
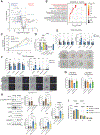
Cancers invoke various pathways to mitigate external and internal stresses to continue their growth and progression. We previously reported that the eIF2 kinase GCN2 and the integrated stress response are constitutively active in prostate cancer (PCa) and are required to maintain amino acid homeostasis needed to fuel tumor growth. However, although loss of GCN2 function reduces intracellular amino acid availability and PCa growth, there is no appreciable cell death. Here, we discovered that the loss of GCN2 in PCa induces prosenescent p53 signaling. This p53 activation occurred through GCN2 inhibition-dependent reductions in purine nucleotides that impaired ribosome biogenesis and, consequently, induced the impaired ribosome biogenesis checkpoint. p53 signaling induced cell cycle arrest and senescence that promoted the survival of GCN2-deficient PCa cells. Depletion of GCN2 combined with loss of p53 or pharmacological inhibition of de novo purine biosynthesis reduced proliferation and enhanced cell death in PCa cell lines, organoids, and xenograft models. Our findings highlight the coordinated interplay between GCN2 and p53 regulation during nutrient stress and provide insight into how they could be targeted in developing new therapeutic strategies for PCa.
Conflict of interest statement
RCW is a member of the advisory board of HiberCell, Inc. KAS is a consultant for HiberCell, Inc. and Aclaris Therapeutics and receives research support from HiberCell, Inc. TGA consults for HiberCell, Inc.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

