Morphoregulatory ADD3 underlies glioblastoma growth and formation of tumor-tumor connections
- PMID: 39592188
- PMCID: PMC11599137
- DOI: 10.26508/lsa.202402823
Morphoregulatory ADD3 underlies glioblastoma growth and formation of tumor-tumor connections
Abstract
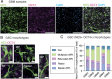
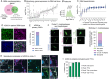
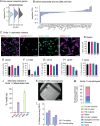
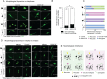
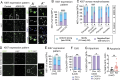
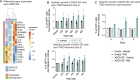
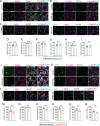
Glioblastoma is a major unmet clinical need characterized by striking inter- and intra-tumoral heterogeneity and a population of glioblastoma stem cells (GSCs), conferring aggressiveness and therapy resistance. GSCs communicate through a network of tumor-tumor connections (TTCs), including nanotubes and microtubes, promoting tumor progression. However, very little is known about the mechanisms underlying TTC formation and overall GSC morphology. As GSCs closely resemble neural progenitor cells during neurodevelopment, we hypothesized that GSCs' morphological features affect tumor progression. We identified GSC morphology as a new layer of tumoral heterogeneity with important consequences on GSC proliferation. Strikingly, we showed that the neurodevelopmental morphoregulator ADD3 is sufficient and necessary for maintaining proper GSC morphology, TTC abundance, cell cycle progression, and chemoresistance, as well as required for cell survival. Remarkably, both the effects on cell morphology and proliferation depend on the stability of actin cytoskeleton. Hence, cell morphology and its regulators play a key role in tumor progression by mediating cell-cell communication. We thus propose that GSC morphological heterogeneity holds the potential to identify new therapeutic targets and diagnostic markers.
© 2024 Barelli et al.
Conflict of interest statement
F Iorio receives funds from Open Targets, a public–private initiative involving academia, and from Nerviano Medical Sciences S.r.l and performs consultancy for the Cancer Research Horizons-AstraZeneca Functional Genomics Centre and for Mosaic Therapeutics.
Figures













References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
