Integrated analyses of multi-omic data derived from paired primary lung cancer and brain metastasis reveal the metabolic vulnerability as a novel therapeutic target
- PMID: 39593114
- PMCID: PMC11590298
- DOI: 10.1186/s13073-024-01410-8
Integrated analyses of multi-omic data derived from paired primary lung cancer and brain metastasis reveal the metabolic vulnerability as a novel therapeutic target
Abstract
Background: Lung cancer brain metastases (LC-BrMs) are frequently associated with dismal mortality rates in patients with lung cancer; however, standard of care therapies for LC-BrMs are still limited in their efficacy. A deep understanding of molecular mechanisms and tumor microenvironment of LC-BrMs will provide us with new insights into developing novel therapeutics for treating patients with LC-BrMs.
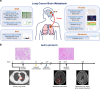
Methods: Here, we performed integrated analyses of genomic, transcriptomic, proteomic, metabolomic, and single-cell RNA sequencing data which were derived from a total number of 154 patients with paired and unpaired primary lung cancer and LC-BrM, spanning four published and two newly generated patient cohorts on both bulk and single cell levels.
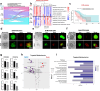
Results: We uncovered that LC-BrMs exhibited a significantly greater intra-tumor heterogeneity. We also observed that mutations in a subset of genes were almost always shared by both primary lung cancers and LC-BrM lesions, including TTN, TP53, MUC16, LRP1B, RYR2, and EGFR. In addition, the genome-wide landscape of somatic copy number alterations was similar between primary lung cancers and LC-BrM lesions. Nevertheless, several regions of focal amplification were significantly enriched in LC-BrMs, including 5p15.33 and 20q13.33. Intriguingly, integrated analyses of transcriptomic, proteomic, and metabolomic data revealed mitochondrial-specific metabolism was activated but tumor immune microenvironment was suppressed in LC-BrMs. Subsequently, we validated our results by conducting real-time quantitative reverse transcription PCR experiments, immunohistochemistry, and multiplexed immunofluorescence staining of patients' paired tumor specimens. Therapeutically, targeting oxidative phosphorylation with gamitrinib in patient-derived organoids of LC-BrMs induced apoptosis and inhibited cell proliferation. The combination of gamitrinib plus anti-PD-1 immunotherapy significantly improved survival of mice bearing LC-BrMs. Patients with a higher expression of mitochondrial metabolism genes but a lower expression of immune genes in their LC-BrM lesions tended to have a worse survival outcome.
Conclusions: In conclusion, our findings not only provide comprehensive and integrated perspectives of molecular underpinnings of LC-BrMs but also contribute to the development of a potential, rationale-based combinatorial therapeutic strategy with the goal of translating it into clinical trials for patients with LC-BrMs.
Keywords: Lung cancer brain metastases (LC-BrMs); Mitochondrial-specific metabolism; Tumor immune microenvironment.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The lung tumor, brain metastasis tissues, and blood samples were derived from patients who were presented at SYSUCC (n = 67) and WCH (n = 5). Patient samples were collected under the Institutional Review Board (IRB) protocols of SYSUCC (Protocol B2021-256–01) and WCH (2019–57), approved by the Medical Ethics Committee of SYSUCC and the Biomedical Ethics Committee of WCH (Sichuan University), respectively. Written informed consent were obtained from all patients. This study was conducted in accordance with the principles of the Helsinki Declaration. Animal. This study, which involved animal models, was approved by the Animal Care and Use Committee of Laboratory Animal Ethics Committee of Affiliated First Hospital of Guangzhou Medical University (Reference No. 2021169), in full compliance with all applicable ethical standards for animal research. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. 10.3322/caac.21660. - PubMed
-
- Riihimaki M, Hemminki A, Fallah M, Thomsen H, Sundquist K, Sundquist J, Hemminki K. Metastatic sites and survival in lung cancer. Lung Cancer. 2014;86:78–84. 10.1016/j.lungcan.2014.07.020. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. 10.3322/caac.21442. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. 10.3322/caac.21387. - PubMed
-
- Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, Zhou C, Hu CP, O’Byrne K, Feng J, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16:141–51. 10.1016/S1470-2045(14)71173-8. - PubMed
MeSH terms
Substances
Grants and funding
- 82303805/National Natural Science Foundation of China
- 82203574/National Natural Science Foundation of China
- 31771549/National Natural Science Foundation of China
- 81872324/National Natural Science Foundation of China
- 2020A1515110069/GuangDong Basic and Applied Basic Research Foundation
- 23ZDYF2661/Science and Technology Support Program of Sichuan Province
- 2022YFS0375/Science and Technology Support Program of Sichuan Province
- 20231800936072/Dongguan Social Development Science and Technology Project
- 2023NSFSC1884/Natural Science Foundation of Sichuan Province
- 2023HXBH105/Postdoctor Research Fund of West China Hospital, Sichuan University
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

