Multidrug-resistant ESBL-producing Klebsiella pneumoniae complex in Czech hospitals, wastewaters and surface waters
- PMID: 39593189
- PMCID: PMC11590221
- DOI: 10.1186/s13756-024-01496-0
Multidrug-resistant ESBL-producing Klebsiella pneumoniae complex in Czech hospitals, wastewaters and surface waters
Erratum in
-
Correction: Multidrug-resistant ESBL-producing Klebsiella pneumoniae complex in Czech hospitals, wastewaters and surface waters.Antimicrob Resist Infect Control. 2024 Dec 16;13(1):146. doi: 10.1186/s13756-024-01508-z. Antimicrob Resist Infect Control. 2024. PMID: 39681860 Free PMC article. No abstract available.
Abstract
Background: Multidrug-resistant (MDR) bacteria pose a significant challenge to the treatment of infectious diseases. Of particular concern are members of the Klebsiella pneumoniae species complex (KpSC), which are frequently associated with hospital-acquired infections and have the potential to spread outside hospitals via wastewaters. In this study, we aimed to investigate the occurrence and phylogenetic relatedness of MDR KpSC from patients with urinary tract infections (UTIs), hospital sewage, municipal wastewater treatment plants (mWWTPs) and surface waters and to evaluate the clinical relevance of the KpSC subspecies.
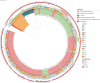
Methods: A total of 372 KpSC isolates resistant to third-generation cephalosporins and/or meropenem were collected from patients (n = 130), hospital sewage (n = 95), inflow (n = 54) and outflow from the mWWTPs (n = 63), river upstream (n = 13) and downstream mWWTPs (n = 17) from three cities in the Czech Republic. The isolates were characterized by antimicrobial susceptibility testing and whole-genome sequencing (Illumina). The presence of antibiotic resistance genes, plasmid replicons and virulence-associated factors was determined. A phylogenetic tree and single nucleotide polymorphism matrix were created to reveal the relatedness between isolates.
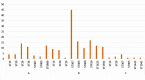
Results: The presence of MDR KpSC isolates (95%) was identified in all water sources and locations. Most isolates (99.7%) produced extended-spectrum beta-lactamases encoded by blaCTX-M-15. Resistance to carbapenems (5%) was observed mostly in wastewaters, but carbapenemase genes, such as blaGES-51 (n = 10), blaOXA-48 (n = 4), blaNDM-1 (n = 4) and blaKPC-3 (n = 1), were found in isolates from all tested locations and different sources except rivers. Among the 73 different sequence types (STs), phylogenetically related isolates were observed only among the ST307 lineage. Phylogenetic analysis revealed the transmission of this lineage from patients to the mWWTP and from the mWWTP to the adjacent river and the presence of the ST307 clone in the mWWTP over eight months. We confirmed the frequent abundance of K. pneumoniae (K. pneumoniae sensu stricto and K. pneumoniae subsp. ozaenae) in patients suffering from UTIs. K. variicola isolates formed only a minor proportion of UTIs, and K. quasipneumoniae was not found among UTIs isolates; however, these subspecies were frequently observed in hospital sewage communities during the first sampling period.
Conclusion: This study provides evidence of the transmission and persistence of the ST307 lineage from UTIs isolates via mWWTPs to surface waters. Isolates from UTIs consisted mostly of K. pneumoniae. Other isolates of KpSC were observed in hospital wastewaters, which implies the impact of sources other than UTIs. This study highlights the influence of urban wastewaters on the spread of MDR KpSC to receiving environments.
Keywords: Klebsiella spp. subspecies; bla CTX−M−15; Urinary tract infections; Wastewater treatment plants.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: Not required. Patient consent: Not required. Permission to reproduce material from other sources: Not required. Clinical trial registration: Not required. Competing interests: The authors declare no competing interests.
Figures



References
-
- Aria M, Farajnia S, Khosroshahi SA, Naghilli B, Farajnia H, Sanjari A, Leylabadlo HE, Tanoumand A. OXA-10 and OXA-2 ESBLs among multidrug-resistant Pseudomonas aeruginosa isolates from North West of Iran. Progress Biol Sci. 2017;7(2):191–7. 10.22059/pbs.2020.260927.1307.
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. 10.1089/cmb.2012.0021. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

