A Comparative Study of Methyl-BEAMing and Droplet Digital PCR for MGMT Gene Promoter Hypermethylation Detection
- PMID: 39594133
- PMCID: PMC11592929
- DOI: 10.3390/diagnostics14222467
A Comparative Study of Methyl-BEAMing and Droplet Digital PCR for MGMT Gene Promoter Hypermethylation Detection
Abstract
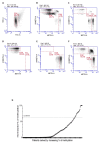
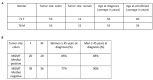
Background: O-6-methylguanine-DNA methyltransferase is responsible for the direct repair of O6-methylguanine lesions induced by alkylating agents, including temozolomide. O-6-methylguanine-DNA methyltransferase promoter hypermethylation is a well-established biomarker for temozolomide response in glioblastoma patients, also correlated with therapeutic response in colorectal cancer. Objectives: The ARETHUSA clinical trial aims to stratify colorectal cancer patients based on their mismatch repair status. Mismatch repair-deficient patients are eligible for treatment with immune checkpoint inhibitors (anti-PDL-1), whereas mismatch repair-proficient samples are screened for O-6-methylguanine-DNA methyltransferase promoter methylation to identify those suitable for temozolomide treatment. Methods: In this context, a subset of ARETHUSA metastatic colorectal cancer samples was used to compare two different techniques for assessing O-6-methylguanine-DNA methyltransferase hypermethylation: Methyl-BEAMing, a highly sensitive digital PCR approach that combines emulsion PCR and flow cytometry, and droplet digital PCR, a more automated procedure that enables the rapid, operator-independent analysis of a large number of samples. Results: Our study clearly demonstrates that the results obtained using Methyl-BEAMing and droplet digital PCR are comparable, with both techniques showing similar accuracy, sensitivity, and reproducibility. Conclusions: Digital droplet PCR proved to be an efficient method for detecting gene promoter methylation. However, the Methyl-BEAMing method has proved more sensitive for detecting low quantities of DNA.
Keywords: DNAmethylation; MGMT; Methyl-BEAMing; digital PCR; metastatic colorectal cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wick W., Gorlia T., Bady P., Platten M., van den Bent M.J., Taphoorn M.J., Steuve J., Brandes A.A., Hamou M.F., Wick A., et al. Phase II Study of Radiotherapy and Temsirolimus versus Radiochemotherapy with Temozolomide in Patients with Newly Diagnosed Glioblastoma without MGMT Promoter Hypermethylation (EORTC 26082) Clin. Cancer Res. 2016;22:4797–4806. doi: 10.1158/1078-0432.CCR-15-3153. - DOI - PubMed
-
- Pietrantonio F., Perrone F., de Braud F., Castano A., Maggi C., Bossi I., Gevorgyan A., Biondani P., Pacifici M., Busico A., et al. Activity of temozolomide in patients with advanced chemorefractory colorectal cancer and MGMT promoter methylation. Ann. Oncol. 2014;25:404–408. doi: 10.1093/annonc/mdt547. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

