Detection of Brain-Derived Cell-Free DNA in Plasma
- PMID: 39594207
- PMCID: PMC11592591
- DOI: 10.3390/diagnostics14222541
Detection of Brain-Derived Cell-Free DNA in Plasma
Abstract
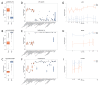
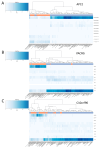
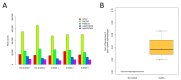
Background: Neuronal loss is a major pathological feature of neurodegenerative diseases. The analysis of plasma cell-free DNA (cfDNA) is an emerging approach to track cell death events in a minimally invasive way and from inaccessible areas of the body, such as the brain. Previous studies showed that DNA methylation (DNAm) profiles can be used to map the tissue of origin of cfDNA and to identify molecules released from the brain upon cell death. The aim of the present study is to contribute to this research field, presenting the development and validation of an assay for the detection of brain-derived cfDNA (bcfDNA). Methods: To identify CpG sites with brain-specific DNAm, we compared brain and non-brain tissues for their chromatin state profiles and genome-wide DNAm data, available in public datasets. The selected target genomic regions were experimentally validated by bisulfite sequencing on DNA extracted from 44 different autoptic tissues, including multiple brain regions. Sequencing data were analysed to identify brain-specific epihaplotypes. The developed assay was tested in plasma cfDNA from patients with immune effector cell-associated neurotoxicity syndrome (ICANS) following chimeric antigen receptor T (CAR-T) therapy. Results: We validated five genomic regions with brain-specific DNAm (four hypomethylated and one hypermethylated in the brain). DNAm analysis of the selected genomic regions in plasma samples from CAR-T patients revealed higher levels of bcfDNA in participants with ongoing neurotoxicity syndrome. Conclusions: We developed an assay for the analysis of bcfDNA in plasma. The assay is a promising tool for the early detection of neuronal loss in neurodegenerative diseases.
Keywords: DNA methylation; cfDNA; epihaplotypes; neurodegeneration; target bisulfite sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Fernández-Lázaro D., García Hernández J.L., García A.C., Córdova Martínez A., Mielgo-Ayuso J., Cruz-Hernández J.J. Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer’s Biomarkers. Diagnostics. 2020;10:215. doi: 10.3390/diagnostics10040215. - DOI - PMC - PubMed
-
- Grabuschnig S., Bronkhorst A.J., Holdenrieder S., Rosales Rodriguez I., Schliep K.P., Schwendenwein D., Ungerer V., Sensen C.W. Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. Int. J. Mol. Sci. 2020;21:8062. doi: 10.3390/ijms21218062. - DOI - PMC - PubMed
Grants and funding
- "Radiogenomics for Early detection of Alzheimer's Disease: combining DNA methylation-based liquid biopsy and advanced Magnetic Resonance Imaging" (READy) (Grant AGYR 2020)/Associazione Italiana Ricerca Alzheimer Onlus (Airalzh)
- project MNESYS (PE0000006)-"A multiscale integrated approach to the study of the nervous sys-tem in health and disease" (DN. 1553 11.10.2022)/Ministry of University and Research (MUR), the National Recovery and Resilience Plan (PNRR)
- project PE00000019, CUP F33C22000500006/MUR under PNRR M4C2I1.3 Heal Italia
- RC-2022-2773291 project "Identificazione di biomarcatori di rispostaclinica e di insorgenza di complicanze in pazientiematologici sottoposti a terapia CAR-T"/Italian Ministry of Health
LinkOut - more resources
Full Text Sources

