Characterization of the Peroxisomal Proteome and Redox Balance in Human Prostate Cancer Cell Lines
- PMID: 39594482
- PMCID: PMC11591464
- DOI: 10.3390/antiox13111340
Characterization of the Peroxisomal Proteome and Redox Balance in Human Prostate Cancer Cell Lines
Abstract
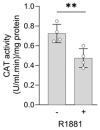
Prostate cancer (PCa) is associated with disruptions in cellular redox balance. Given the intricate role of peroxisomes in redox metabolism, we conducted comprehensive proteomics analyses to compare peroxisomal and redox protein profiles between benign (RWPE-1) and malignant (22Rv1, LNCaP, and PC3) prostate cell lines. Our analyses revealed significant enrichment of the "peroxisome" pathway among proteins notably upregulated in androgen receptor (AR)-positive cell lines. In addition, catalase (CAT) activity was consistently higher in these malignant cell lines compared to RWPE-1, which contrasts with previous studies reporting lower CAT levels and increased H2O2 levels in PCa tissues compared to adjacent normal tissues. To mimic this clinical scenario, we used RNA interference to knock down CAT expression. Our results show that reduced CAT levels enhanced 22Rv1 and LNCaP cell proliferation. R1881-induced activation of AR, a key driver of PCa, increased expression of the H2O2-producing peroxisomal β-oxidation enzymes acyl-coenzyme A oxidase 1 and 3, reduced CAT expression and activity, and elevated peroxisomal H2O2 levels. Considering these changes and other antioxidant enzyme profile alterations, we propose that enhanced AR activity in PCa reduces CAT function, leading to increased peroxisomal H2O2 levels that trigger adaptive stress responses to promote cell survival, growth, and proliferation.
Keywords: R1881; androgen receptor; catalase; hydrogen peroxide; peroxisomes; prostate cancer.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of this study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures











References
-
- Global Cancer Observatory: Cancer Today (Version 1.1); International Agency for Research on Cancer: Lyon, France. [(accessed on 9 September 2024)]. Available online: https://gco.iarc.who.int/today.
-
- Battisti V., Maders L.D., Bagatini M.D., Reetz L.G., Chiesa J., Battisti I.E., Gonçalves J.F., Duarte M.M., Schetinger M.R., Morsch V.M. Oxidative stress and antioxidant status in prostate cancer patients: Relation to Gleason score, treatment and bone metastasis. Biomed. Pharmacother. 2011;65:516–524. doi: 10.1016/j.biopha.2011.06.003. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

