LKB1 and STRADα Promote Epithelial Ovarian Cancer Spheroid Cell Invasion
- PMID: 39594681
- PMCID: PMC11591840
- DOI: 10.3390/cancers16223726
LKB1 and STRADα Promote Epithelial Ovarian Cancer Spheroid Cell Invasion
Abstract
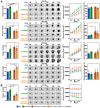
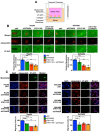
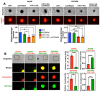
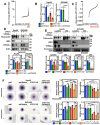
Late-stage epithelial ovarian cancer (EOC) involves the widespread dissemination of malignant disease throughout the peritoneal cavity, often accompanied by ascites. EOC metastasis relies on the formation of multicellular aggregates, called spheroids. Given that Liver Kinase B1 (LKB1) is required for EOC spheroid viability and LKB1 loss in EOC cells decreases tumor burden in mice, we investigated whether the LKB1 complex controls the invasive properties of human EOC spheroids. LKB1 signalling was antagonized through the CRISPR/Cas9 genetic knockout of LKB1 and/or the RNAi-dependent targeting of STE20-related kinase adaptor protein (STRAD, an LKB1 activator). EOC spheroids expressing nuclear GFP (green) or mKate2 (red) constructs were embedded in Matrigel for real-time live-cell invasion monitoring. Migration and invasion were also assessed in spheroid culture using Transwell chambers, spheroid reattachment, and mesothelial clearance assays. The loss of LKB1 and STRAD signalling decreased cell invasion through Matrigel and Transwell membranes, as well as mesothelial cell clearance. In the absence of LKB1, zymographic assays identified a loss of matrix metalloproteinase (MMP) activity, whereas spheroid reattachment assays found that coating plates with fibronectin restored their invasive potential. A three-dimensional EOC organoid model demonstrated that organoid area was greatly reduced by LKB1 loss. Overall, our data indicated that LKB1 and STRAD facilitated EOC metastasis by promoting MMP activity and fibronectin expression. Given that LKB1 and STRAD are crucial for EOC metastasis, targeting LKB1 and/or STRAD could disrupt the dissemination of EOC, making inhibitors of the LKB1 pathway an alternative therapeutic strategy for EOC patients.
Keywords: STE20-related kinase adaptor protein (STRAD); epithelial ovarian cancer (EOC); liver kinase B1 (LKB1); mesothelial clearance; metastasis; organoid; spheroid.
Conflict of interest statement
The authors declare that they have no affiliations with or involvement in any organization or entity with any financial interest in the subject matter or materials discussed in this manuscript.
Figures



References
-
- Menon U., Gentry-Maharaj A., Burnell M., Singh N., Ryan A., Karpinskyj C., Carlino G., Taylor J., Massingham S.K., Raikou M., et al. Ovarian cancer population screening and mortality after long-term follow-up in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): A randomised controlled trial. Lancet. 2021;397:2182–2193. doi: 10.1016/S0140-6736(21)00731-5. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

