Exploring Extracellular Vesicle Surface Protein Markers Produced by Glioblastoma Tumors: A Characterization Study Using In Vitro 3D Patient-Derived Cultures
- PMID: 39594703
- PMCID: PMC11592176
- DOI: 10.3390/cancers16223748
Exploring Extracellular Vesicle Surface Protein Markers Produced by Glioblastoma Tumors: A Characterization Study Using In Vitro 3D Patient-Derived Cultures
Abstract
Background/objectives: Glioblastoma (GBM) is an aggressive brain cancer with limited treatment options. Extracellular vesicles (EVs) derived from GBM cells contain important biomarkers, such as microRNAs, proteins, and DNA mutations, which are involved in tumor progression, invasion, and resistance to treatment. Identifying surface markers on these EVs is crucial for their isolation and potential use in noninvasive diagnosis. This study aimed to use tumor-derived explants to investigate the surface markers of EVs and explore their role as diagnostic biomarkers for GBM.
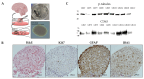
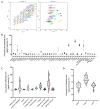
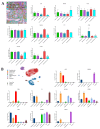
Methods: Tumor explants from nine GBM patients without IDH1/IDH2 mutations or 1p-19q co-deletion were cultured to preserve both tumor viability and cytoarchitecture. EVs were collected from the tumor microenvironment using differential centrifugation, filtration, and membrane affinity binding. Their surface protein composition was analyzed through multiplex protein assays. RNA-Seq data from TCGA and GTEx datasets, along with in silico single-cell RNA-seq data, were used to assess EV surface biomarker expression across large GBM patient cohorts.
Results: The in vitro model successfully replicated the tumor microenvironment and produced EVs with distinct surface markers. Biomarker analysis in large datasets revealed specific expression patterns unique to GBM patients compared with healthy controls. These markers demonstrated potential as a GBM-specific signature and were correlated with clinical data. Furthermore, in silico single-cell RNA-seq provided detailed insights into biomarker distribution across different cell types within the tumor.
Conclusions: This study underscores the efficacy of the tumor-derived explant model and its potential to advance the understanding of GBM biology and EV production. A key innovation is the isolation of EVs from a model that faithfully mimics the tumor's original cytoarchitecture, offering a deeper understanding of the cells involved in EV release. The identified EV surface markers represent promising targets for enhancing EV isolation and optimizing their use as diagnostic tools. Moreover, further investigation into their molecular cargo may provide crucial insights into tumor characteristics and evolution.
Keywords: extracellular vesicles; glioblastoma; surface markers; tumor microenvironment; tumor-derived explants.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Nikolova E., Laleva L., Milev M., Spiriev T., Stoyanov S., Ferdinandov D., Mitev V., Todorova A. miRNAs and related genetic biomarkers according to the WHO glioma classification: From diagnosis to future therapeutic targets. Non-Coding RNA Res. 2023;9:141–152. doi: 10.1016/j.ncrna.2023.10.003. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

