Metabolomic Profiling and Machine Learning Models for Tumor Classification in Patients with Recurrent IDH-Wild-Type Glioblastoma: A Prospective Study
- PMID: 39594811
- PMCID: PMC11593314
- DOI: 10.3390/cancers16223856
Metabolomic Profiling and Machine Learning Models for Tumor Classification in Patients with Recurrent IDH-Wild-Type Glioblastoma: A Prospective Study
Abstract
Background/objectives: The recurrence of glioblastoma is an inevitable event in this disease's course. In this study, we sought to identify the metabolomic signature in patients with recurrent glioblastomas undergoing surgery and radiation therapy.
Methods: Blood samples collected prospectively from six patients with recurrent IDH-wildtype glioblastoma who underwent one surgery at diagnosis and a second surgery at relapse were analyzed using untargeted gas chromatography-time-of-flight mass spectrometry to measure metabolite abundance. The data analysis techniques included univariate analysis, correlation analysis, and a sample t-test. For predictive modeling, machine learning (ML) algorithms such as multinomial logistic regression, gradient boosting, and random forest were applied to predict the classification of samples in the correct treatment phase.
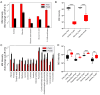
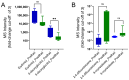
Results: Comparing samples after the first surgery and after the relapse surgeries to the pre-operative samples showed a significant decrease in sorbitol and mannitol; there was a significant increase in urea, oxoproline, glucose, and alanine. After chemoradiation, two metabolites, erythritol and 6-deoxyglucitol, showed a decrease, with a cut-off of three and a significant reduction for 6-deoxyglucitol, while 2,4-difluorotoluene and 9-myristoleate showed an increase post radiation, with a fold-change cut-off of three. The gradient-boosting ML model achieved a high performance for the prediction of tumor conditions in patients with glioblastoma who had undergone relapse surgery.
Conclusions: We developed an ML predictor for tumor phase based on the plasma metabolomic profile. Our study suggests the potential of combining metabolomics with ML as a new tool to stratify the risk of tumor progression in patients with glioblastoma.
Keywords: glioblastoma; machine learning; metabolomics; recurrence.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Application of Machine Learning to Metabolomic Profile Characterization in Glioblastoma Patients Undergoing Concurrent Chemoradiation.Metabolites. 2023 Feb 17;13(2):299. doi: 10.3390/metabo13020299. Metabolites. 2023. PMID: 36837918 Free PMC article.
-
Profile Characterization of Biogenic Amines in Glioblastoma Patients Undergoing Standard-of-Care Treatment.Biomedicines. 2023 Aug 13;11(8):2261. doi: 10.3390/biomedicines11082261. Biomedicines. 2023. PMID: 37626757 Free PMC article.
-
Using Machine Learning to Identify Metabolomic Signatures of Pediatric Chronic Kidney Disease Etiology.J Am Soc Nephrol. 2022 Feb;33(2):375-386. doi: 10.1681/ASN.2021040538. Epub 2022 Jan 11. J Am Soc Nephrol. 2022. PMID: 35017168 Free PMC article.
-
Congress of neurological surgeons systematic review and evidence-based guidelines update on the role of emerging developments in the management of newly diagnosed glioblastoma.J Neurooncol. 2020 Nov;150(2):269-359. doi: 10.1007/s11060-020-03607-4. Epub 2020 Nov 19. J Neurooncol. 2020. PMID: 33215345
-
Advanced Magnetic Resonance Imaging in Pediatric Glioblastomas.Front Neurol. 2021 Nov 10;12:733323. doi: 10.3389/fneur.2021.733323. eCollection 2021. Front Neurol. 2021. PMID: 34858308 Free PMC article. Review.
Cited by
-
Towards non-invasive diagnosis of glioblastoma: identifying metabolic biomarkers in liquid biopsies using a ROC-based approach.Discov Oncol. 2025 Aug 2;16(1):1456. doi: 10.1007/s12672-025-03310-8. Discov Oncol. 2025. PMID: 40751888 Free PMC article.
References
-
- Hertler C., Felsberg J., Gramatzki D., Le Rhun E., Clarke J., Soffietti R., Wick W., Chinot O., Ducray F., Roth P., et al. Long-term survival with idh wildtype glioblastoma: First results from the eternity brain tumor funders’ collaborative consortium (eortc 1419) Eur. J. Cancer. 2023;189:112913. doi: 10.1016/j.ejca.2023.05.002. - DOI - PubMed
-
- Louis D.N., Perry A., Wesseling P., Brat D.J., Cree I.A., Figarella-Branger D., Hawkins C., Ng H.K., Pfister S.M., Reifenberger G., et al. The 2021 who classification of tumors of the central nervous system: A summary. Neuro-Oncology. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. - DOI - PMC - PubMed
-
- Yoo J., Yoon S.J., Kim K.H., Jung I.H., Lim S.H., Kim W., Yoon H.I., Kim S.H., Sung K.S., Roh T.H., et al. Patterns of recurrence according to the extent of resection in patients with idh-wild-type glioblastoma: A retrospective study. J. Neurosurg. 2021;137:533–543. doi: 10.3171/2021.10.JNS211491. - DOI - PubMed
-
- Jacob F., Salinas R.D., Zhang D.Y., Nguyen P.T.T., Schnoll J.G., Wong S.Z.H., Thokala R., Sheikh S., Saxena D., Prokop S., et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204.e22. doi: 10.1016/j.cell.2019.11.036. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

