Restored Collagen VI Microfilaments Network in the Extracellular Matrix of CRISPR-Edited Ullrich Congenital Muscular Dystrophy Fibroblasts
- PMID: 39595588
- PMCID: PMC11591638
- DOI: 10.3390/biom14111412
Restored Collagen VI Microfilaments Network in the Extracellular Matrix of CRISPR-Edited Ullrich Congenital Muscular Dystrophy Fibroblasts
Abstract
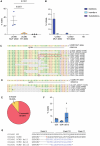
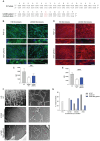
Collagen VI is an essential component of the extracellular matrix (ECM) composed by α1, α2 and α3 chains and encoded by COL6A1, COL6A2 and COL6A3 genes. Dominant negative pathogenic variants in COL6A genes result in defects in collagen VI protein and are implicated in the pathogenesis of muscular diseases, including Ullrich congenital muscular dystrophy (UCMD). Here, we designed a CRISPR genome editing strategy to tackle a dominant heterozygous deletion c.824_838del in exon 9 of the COL6A1 gene, causing a lack of secreted collagen VI in a patient's dermal fibroblasts. The evaluation of efficiency and specificity of gene editing in treating patient's fibroblasts revealed the 32% efficiency of editing the mutated allele but negligible editing of the wild-type allele. CRISPR-treated UCMD skin fibroblasts rescued the secretion of collagen VI in the ECM, which restored the ultrastructure of the collagen VI microfibril network. By using normal melanocytes as surrogates of muscle cells, we found that collagen VI secreted by the corrected patient's skin fibroblasts recovered the anchorage to the cell surface, pointing to a functional improvement of the protein properties. These results support the application of the CRISPR editing approach to knock out COL6A1 mutated alleles and rescue the UCMD phenotype in patient-derived fibroblasts.
Keywords: CRISPR/Cas9; allele-specific gene editing; collagen VI; collagen VI-related disorders; patient-derived fibroblasts.
Conflict of interest statement
The authors declare no conflicts of interests.
Figures



References
-
- Camacho Vanegas O., Bertini E., Zhang R.Z., Petrini S., Minosse C., Sabatelli P., Giusti B., Chu M.L., Pepe G. Ullrich scleroatonic muscular dystrophy is caused by recessive mutations in collagen type VI. Proc. Natl. Acad. Sci. USA. 2001;98:7516–7521. doi: 10.1073/pnas.121027598. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

