Mechanistic and Therapeutic Implications of Protein and Lipid Sialylation in Human Diseases
- PMID: 39596031
- PMCID: PMC11594235
- DOI: 10.3390/ijms252211962
Mechanistic and Therapeutic Implications of Protein and Lipid Sialylation in Human Diseases
Abstract
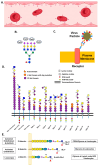
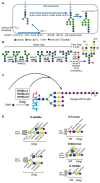
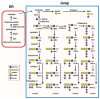
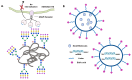
Glycan structures of glycoproteins and glycolipids on the surface glycocalyx and luminal sugar layers of intracellular membrane compartments in human cells constitute a key interface between intracellular biological processes and external environments. Sialic acids, a class of alpha-keto acid sugars with a nine-carbon backbone, are frequently found as the terminal residues of these glycoconjugates, forming the critical components of these sugar layers. Changes in the status and content of cellular sialic acids are closely linked to many human diseases such as cancer, cardiovascular, neurological, inflammatory, infectious, and lysosomal storage diseases. The molecular machineries responsible for the biosynthesis of the sialylated glycans, along with their biological interacting partners, are important therapeutic strategies and targets for drug development. The purpose of this article is to comprehensively review the recent literature and provide new scientific insights into the mechanisms and therapeutic implications of sialylation in glycoproteins and glycolipids across various human diseases. Recent advances in the clinical developments of sialic acid-related therapies are also summarized and discussed.
Keywords: Siglecs and Selectins; clinical development of therapies; gangliosides; glycocalyx; human diseases; sialylation of glycoproteins and glycolipids.
Conflict of interest statement
All authors are employed by Pfizer. All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures

References
-
- Blix G.S., Svennerholm L., Werner I., Finsnes E., Sorensen J.S., Sorensen N.A. The Isolation of Chondrosamine from Gangliosides and from Submaxillary Mucin. Acta Chem. Scand. 1952;6:358–362. doi: 10.3891/acta.chem.scand.06-0358. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

