Transcriptomic Changes During the Replicative Senescence of Human Articular Chondrocytes
- PMID: 39596199
- PMCID: PMC11594096
- DOI: 10.3390/ijms252212130
Transcriptomic Changes During the Replicative Senescence of Human Articular Chondrocytes
Abstract
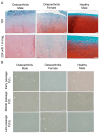
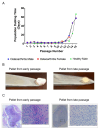
Aging is a major risk factor for osteoarthritis (OA), but the specific mechanisms connecting aging and OA remain unclear. Although chondrocytes rarely divide in adult articular cartilage, they undergo replicative senescence in vitro, offering a model to study aging-related changes under controlled conditions. OA cartilage was obtained from an 80-year-old male and a 72-year-old female, while normal cartilage was sourced from a 26-year-old male. Chondrocyte cultures were established and sub-cultured to their Hayflick limit. Bulk RNA sequencing on early- and late-passage human articular chondrocytes identified transcriptomic changes associated with cellular aging. Early-passage OA chondrocytes already showed senescent phenotypes, unlike normal chondrocytes. All three cultures underwent 30 population doublings before replicative exhaustion, at which point all cells displayed senescence. During this process, cells lost their ability to form cartilaginous pellets. Differential gene expression analysis revealed distinct transcriptomic profiles between early- and late-passage chondrocytes and between normal and OA-derived cells. Genes related to matrix synthesis, degradation, inflammation, and the senescence-associated secretory phenotype (SASP) showed significant expression changes. Despite being a small pilot study, these findings suggest that further research into the molecular and metabolic changes during chondrocyte senescence could provide valuable insights into OA pathobiology.
Keywords: Hayflick limit; aging; chondrocyte replicative senescence; osteoarthritis; transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







Update of
-
Transcriptomic changes during the replicative senescence of human articular chondrocytes.bioRxiv [Preprint]. 2023 Nov 7:2023.11.07.565835. doi: 10.1101/2023.11.07.565835. bioRxiv. 2023. Update in: Int J Mol Sci. 2024 Nov 12;25(22):12130. doi: 10.3390/ijms252212130. PMID: 37986862 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

