Single-Sample Networks Reveal Intra-Cytoband Co-Expression Hotspots in Breast Cancer Subtypes
- PMID: 39596229
- PMCID: PMC11594411
- DOI: 10.3390/ijms252212163
Single-Sample Networks Reveal Intra-Cytoband Co-Expression Hotspots in Breast Cancer Subtypes
Abstract
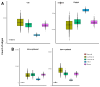
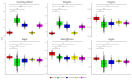
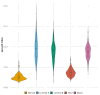
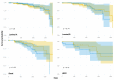
Breast cancer is a heterogeneous disease comprising various subtypes with distinct molecular characteristics, clinical outcomes, and therapeutic responses. This heterogeneity evidences significant challenges for diagnosis, prognosis, and treatment. Traditional genomic co-expression network analyses often overlook individual-specific interactions critical for personalized medicine. In this study, we employed single-sample gene co-expression network analysis to investigate the structural and functional genomic alterations across breast cancer subtypes (Luminal A, Luminal B, Her2-enriched, and Basal-like) and compared them with normal breast tissue. We utilized RNA-Seq gene expression data to infer gene co-expression networks. The LIONESS algorithm allowed us to construct individual networks for each patient, capturing unique co-expression patterns. We focused on the top 10,000 gene interactions to ensure consistency and robustness in our analysis. Network metrics were calculated to characterize the topological properties of both aggregated and single-sample networks. Our findings reveal significant fragmentation in the co-expression networks of breast cancer subtypes, marked by a change from interchromosomal (TRANS) to intrachromosomal (CIS) interactions. This transition indicates disrupted long-range genomic communication, leading to localized genomic regulation and increased genomic instability. Single-sample analyses confirmed that these patterns are consistent at the individual level, highlighting the molecular heterogeneity of breast cancer. Despite these pronounced alterations, the proportion of CIS interactions did not significantly correlate with patient survival outcomes across subtypes, suggesting limited prognostic value. Furthermore, we identified high-degree genes and critical cytobands specific to each subtype, providing insights into subtype-specific regulatory networks and potential therapeutic targets. These genes play pivotal roles in oncogenic processes and may represent important keys for targeted interventions. The application of single-sample co-expression network analysis proves to be a powerful tool for uncovering individual-specific genomic interactions.
Keywords: breast cancer networks; co-expression networks; intra-chromosomal hotspots; intra-cytoband co-expression; single-sample networks.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

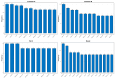
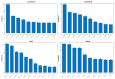
References
-
- Sørlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
-
- Albert R., Barabasi A.L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74:47–97. doi: 10.1103/RevModPhys.74.47. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

