Integrating Physiology, Transcriptome, and Metabolome Analyses Reveals the Drought Response in Two Quinoa Cultivars with Contrasting Drought Tolerance
- PMID: 39596254
- PMCID: PMC11594460
- DOI: 10.3390/ijms252212188
Integrating Physiology, Transcriptome, and Metabolome Analyses Reveals the Drought Response in Two Quinoa Cultivars with Contrasting Drought Tolerance
Abstract
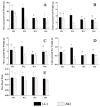
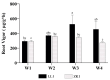
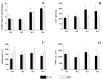
Quinoa (Chenopodium quinoa Willd.) is an annual broadleaf plant belonging to the Amaranthaceae family. It is a nutritious food crop and is considered to be drought-tolerant, but drought is still one of the most important abiotic stress factors limiting its yield. Quinoa responses to drought are related to drought intensity and genotype. This study used two different drought-responsive quinoa cultivars, LL1 (drought-tolerant) and ZK1 (drought-sensitive), to reveal the important mechanisms of drought response in quinoa by combining physiological, transcriptomic, and metabolomic analyses. The physiological analysis indicated that Chla/Chlb might be important for drought tolerance in quinoa. A total of 1756 and 764 differentially expressed genes (DEGs) were identified in LL1 and ZK1, respectively. GO (Gene Ontology) enrichment analysis identified 52 common GO terms, but response to abscisic acid (GO:0009737) and response to osmotic stress (GO:0006970) were only enriched in LL1. KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis revealed that glycerophospholipid metabolism (ko00564) and cysteine and methionine metabolism (ko00270) ranked at the top of the list in both cultivars. A total of 1844 metabolites were identified by metabolomic analysis. "Lipids and lipid-like" molecules had the highest proportions. The DEMs in LL1 and ZK1 were mainly categorized 6 and 4 Human Metabolome Database (HMDB) superclasses, respectively. KEGG analysis revealed that the 'α-linolenic acid metabolism' was enriched in both LL1 and ZK1. Joint KEGG analysis also revealed that the 'α-linolenic acid metabolism' pathway was enriched by both the DEGs and DEMs of LL1. There were 17 DEGs and 8 DEMs enriched in this pathway, and methyl jasmonate (MeJA) may play an important role in the drought response of quinoa. This study will provide information for the identification of drought resistance in quinoa, research on the molecular mechanism of drought resistance, and genetic breeding for drought resistance in quinoa.
Keywords: Chla/Chlb; drought; methyl jasmonate; multi-omics; quinoa; α-linolenic acid metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
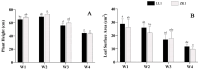











References
-
- Aslam M.U., Raza M.A.S., Saleem M.F., Waqas M., Iqbal R., Ahmad S., Haider I. Improving strategic growth stage-based drought tolerance in quinoa by rhizobacterial inoculation. Commun. Soil Sci. Plant Anal. 2020;51:853–868. doi: 10.1080/00103624.2020.1744634. - DOI
-
- Howell T.A. Enhancing water use efficiency in irrigated agriculture. Agron. J. 2001;93:281–289. doi: 10.2134/agronj2001.932281x. - DOI
-
- Yasui Y., Hirakawa H., Oikawa T., Toyoshima M., Matsuzaki C., Ueno M., Mizuno N., Nagatoshi Y., Imamura T., Miyago M., et al. Draft genome sequence of an inbred line of Chenopodium quinoa, an allotetraploid crop with great environmental adaptability and outstanding nutritional properties. DNA Res. 2016;23:535–546. doi: 10.1093/dnares/dsw037. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

