Relationship Between the Presence of Red Complex Species and the Distribution of Other Oral Bacteria, Including Major Periodontal Pathogens in Older Japanese Individuals
- PMID: 39596308
- PMCID: PMC11594330
- DOI: 10.3390/ijms252212243
Relationship Between the Presence of Red Complex Species and the Distribution of Other Oral Bacteria, Including Major Periodontal Pathogens in Older Japanese Individuals
Abstract
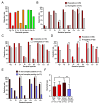
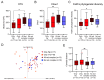
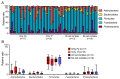
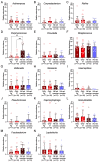
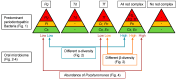
Red complex bacteria (Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia) have high virulence in periodontal disease. In the present study, we aimed to elucidate the detailed symbiotic relationships between the red complex and other oral bacteria in older Japanese individuals. Polymerase chain reaction was performed using dental plaque from 116 subjects and specific primers for ten periodontal pathogens. The detection rate of Prevotella intermedia and Capnocytophaga sputigena was significantly higher in P. gingivalis-positive subjects than in P. gingivalis-negative subjects (p < 0.05). The detection rate of Campylobacter rectus, Prevotella nigrescens, Capnocytophaga ochracea, and Eikenella corrodens was significantly higher in T. forsythia-positive subjects than in T. forsythia-negative subjects (p < 0.01). In a comprehensive analysis of oral microbiomes, three red complex species-positive subjects had significantly higher α-diversity than only P. gingivalis-positive subjects (p < 0.05) and had significantly lower β-diversity than only T. forsythia-positive subjects (p < 0.01). In the taxonomy analysis, Porphyromonas was significantly higher in three red complex species-positive subjects than in only P. gingivalis-positive and only T. forsythia-positive subjects (p < 0.01). These results suggest that each red complex species forms a unique oral microbiome and individuals positive for all red complex bacteria may harbor oral bacteria that confer a significant advantage in developing periodontal disease.
Keywords: Porphyromonas gingivalis; Tannerella forsythia; dental plaque; oral microbiome; red complex species.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Wake N., Asahi Y., Noiri Y., Hayashi M., Motooka D., Nakamura S., Gotoh K., Miura J., Machi H., Iida T., et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. NPJ Biofilms Microbiomes. 2016;2:16018. doi: 10.1038/npjbiofilms.2016.18. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

