A Snapshot of Early Transcriptional Changes Accompanying the Pro-Neural Phenotype Switch by NGN2, ASCL1, SOX2, and MSI1 in Human Fibroblasts: An RNA-Seq Study
- PMID: 39596450
- PMCID: PMC11594342
- DOI: 10.3390/ijms252212385
A Snapshot of Early Transcriptional Changes Accompanying the Pro-Neural Phenotype Switch by NGN2, ASCL1, SOX2, and MSI1 in Human Fibroblasts: An RNA-Seq Study
Abstract
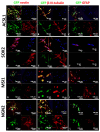
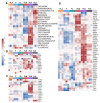
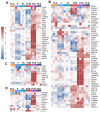
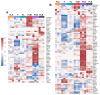
Direct pro-neural reprogramming is a conversion of differentiated somatic cells to neural cells without an intermediate pluripotency stage. It is usually achieved via ectopic expression (EE) of certain transcription factors (TFs) or other reprogramming factors (RFs). Determining the transcriptional changes (TCs) caused by particular RFs in a given cell line enables an informed approach to reprogramming initiation. Here, we characterized TCs in the human fibroblast cell line LF1 on the 5th day after EE of the single well-known pro-neural RFs NGN2, ASCL1, SOX2, and MSI1. As assessed by expression analysis of the bona fide neuronal markers nestin and beta-III tubulin, all four RFs initiated pro-neuronal phenotype conversion; analysis by RNA-seq revealed striking differences in the resulting TCs, although some pathways were overlapping. ASCL1 and SOX2 were not sufficient to induce significant pro-neural phenotype switches using our EE system. NGN2 induced TCs indicative of cell phenotype changes towards neural crest cells, neural stem cells, mature neurons, as well as radial glia, astrocytes, and oligodendrocyte precursors and their mature forms. MSI1 mainly induced a switch towards early stem-like cells, such as radial glia.
Keywords: RNA-seq; cell fate; cell reprogramming; cell therapy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

