Photocleavable Guide crRNAs for a Light-Controllable CRISPR/Cas9 System
- PMID: 39596457
- PMCID: PMC11594570
- DOI: 10.3390/ijms252212392
Photocleavable Guide crRNAs for a Light-Controllable CRISPR/Cas9 System
Abstract
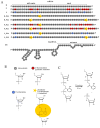
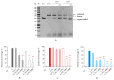
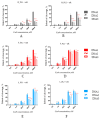
The design of controllable and precise RNA-targeted CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats) systems is an important problem of modern molecular biology and genetic technology. Herein, we have designed a series of photocleavable guide CRISPR RNAs (crRNA) and their 2'-modified (2'-fluoro and locked nucleic acid) analogs containing one or two 1-(2-nitrophenyl)-1,2-ethanediol photolabile linkers (PL). We have demonstrated that these crRNAs can be destroyed by relatively mild UVA irradiation with the rate constants 0.24-0.77 min-1 and that the photocleavage markedly slows down the action of Cas9 nuclease in the model in vitro system. Two PLs provide more rapid crRNA destruction than a single linker. PLs in the crRNA structure improve the specificity of DNA cleavage by Cas9 nuclease for the fully complementary target. The application of photocleavable crRNA in CRISPR/Cas9 genome editing permits the system to be switched off in a spatiotemporally controlled manner, thus alleviating its off-target effects.
Keywords: 2′-modified crRNA; off-target effects; photocleavable crRNA; photoregulated CRISPR/Cas9 system.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

