Genomic Insights into Tibetan Sheep Adaptation to Different Altitude Environments
- PMID: 39596459
- PMCID: PMC11594602
- DOI: 10.3390/ijms252212394
Genomic Insights into Tibetan Sheep Adaptation to Different Altitude Environments
Abstract
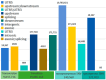
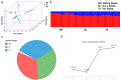
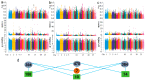
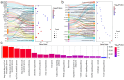
In recent years, research has gradually uncovered the mechanisms of animal adaptation to hypoxic conditions in different altitude environments, particularly at the genomic level. However, past genomic studies on high-altitude adaptation have often not delved deeply into the differences between varying altitude levels. This study conducted whole-genome sequencing on 60 Tibetan sheep (Medium Altitude Group (MA): 20 Tao sheep (TS) at 2887 m, High Altitude Group (HA): 20 OuLa sheep (OL) at 3501 m, and Ultra-High Altitude Group (UA): 20 AWang sheep (AW) at 4643 m) from different regions of the Tibetan Plateau in China to assess their responses under varying conditions. Population genetic structure analysis revealed that the three groups are genetically independent, but the TS and OL groups have experienced gene flow with other northern Chinese sheep due to geographical factors. Selection signal analysis identified FGF10, MMP14, SLC25A51, NDUFB8, ALAS1, PRMT1, PRMT5, and HIF1AN as genes associated with ultra-high-altitude hypoxia adaptation, while HMOX2, SEMA4G, SLC16A2, SLC22A17, and BCL2L2 were linked to high-altitude hypoxia adaptation. Functional analysis showed that ultra-high-altitude adaptation genes tend to influence physiological mechanisms directly affecting oxygen uptake, such as lung development, angiogenesis, and red blood cell formation. In contrast, high-altitude adaptation genes are more inclined to regulate mitochondrial DNA replication, iron homeostasis, and calcium signaling pathways to maintain cellular function. Additionally, the functions of shared genes further support the adaptive capacity of Tibetan sheep across a broad geographic range, indicating that these genes offer significant selective advantages in coping with oxygen scarcity. In summary, this study not only reveals the genetic basis of Tibetan sheep adaptation to different altitudinal conditions but also highlights the differences in gene regulation between ultra-high- and high-altitude adaptations. These findings offer new insights into the adaptive evolution of animals in extreme environments and provide a reference for exploring adaptation mechanisms in other species under hypoxic conditions.
Keywords: Tibetan sheep; high altitude; selection signature; ultra-high altitude; whole-genome resequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Swenson E.R., Bärtsch P. High Altitude. Springer; New York, NY, USA: 2014.
-
- Kappen L. The Lichens. Elsevier; Amsterdam, The Netherlands: 1973. Response to extreme environments; pp. 311–380.
-
- Hu X.-J., Yang J., Xie X.-L., Lv F.-H., Cao Y.-H., Li W.-R., Liu M.-J., Wang Y.-T., Li J.-Q., Liu Y.-G. The genome landscape of Tibetan sheep reveals adaptive introgression from argali and the history of early human settlements on the Qinghai–Tibetan Plateau. Mol. Biol. Evol. 2019;36:283–303. doi: 10.1093/molbev/msy208. - DOI - PMC - PubMed
-
- Beall C.M., Cavalleri G.L., Deng L., Elston R.C., Gao Y., Knight J., Li C., Li J.C., Liang Y., McCormack M. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. - DOI - PMC - PubMed
MeSH terms
Grants and funding
- 2022YFD1302103/National Key R&D Program of China
- 25-LZIHPS-07/Innovation Project of Chinese Academy of Agricultural Sciences
- 22CX8NA014/Science and Technology Program of Gansu Province
- 1610322024012/Central Public-interest Scientific Institution Basal Research Fund
- CARS-39-02/National Technical System for Wool Sheep Industry
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

