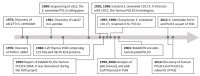
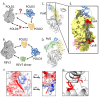
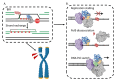
POLD3 as Controller of Replicative DNA Repair
- PMID: 39596481
- PMCID: PMC11595029
- DOI: 10.3390/ijms252212417
POLD3 as Controller of Replicative DNA Repair
Abstract
Multiple modes of DNA repair need DNA synthesis by DNA polymerase enzymes. The eukaryotic B-family DNA polymerase complexes delta (Polδ) and zeta (Polζ) help to repair DNA strand breaks when primed by homologous recombination or single-strand DNA annealing. DNA synthesis by Polδ and Polζ is mutagenic, but is needed for the survival of cells in the presence of DNA strand breaks. The POLD3 subunit of Polδ and Polζ is at the heart of DNA repair by recombination, by modulating polymerase functions and interacting with other DNA repair proteins. We provide the background to POLD3 discovery, investigate its structure, as well as function in cells. We highlight unexplored structural aspects of POLD3 and new biochemical data that will help to understand the pivotal role of POLD3 in DNA repair and mutagenesis in eukaryotes, and its impact on human health.
Keywords: DNA repair; DNA replication; POLD3; Polδ; Polζ; helicase; histone.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
The POLD3 subunit of DNA polymerase δ can promote translesion synthesis independently of DNA polymerase ζ.Nucleic Acids Res. 2015 Feb 18;43(3):1671-83. doi: 10.1093/nar/gkv023. Epub 2015 Jan 27. Nucleic Acids Res. 2015. PMID: 25628356 Free PMC article.
-
Interaction between the Rev1 C-Terminal Domain and the PolD3 Subunit of Polζ Suggests a Mechanism of Polymerase Exchange upon Rev1/Polζ-Dependent Translesion Synthesis.Biochemistry. 2016 Apr 5;55(13):2043-53. doi: 10.1021/acs.biochem.5b01282. Epub 2016 Mar 24. Biochemistry. 2016. PMID: 26982350 Free PMC article.
-
In vivo evidence for translesion synthesis by the replicative DNA polymerase δ.Nucleic Acids Res. 2016 Sep 6;44(15):7242-50. doi: 10.1093/nar/gkw439. Epub 2016 May 16. Nucleic Acids Res. 2016. PMID: 27185888 Free PMC article.
-
POLD1: Central mediator of DNA replication and repair, and implication in cancer and other pathologies.Gene. 2016 Sep 15;590(1):128-41. doi: 10.1016/j.gene.2016.06.031. Epub 2016 Jun 16. Gene. 2016. PMID: 27320729 Free PMC article. Review.
-
Structure and function of eukaryotic DNA polymerase δ.Subcell Biochem. 2012;62:217-36. doi: 10.1007/978-94-007-4572-8_12. Subcell Biochem. 2012. PMID: 22918588 Review.
Cited by
-
Beyond proofreading: POLD1 mutations as dynamic orchestrators of genomic instability and immune evasion in cancer.Front Immunol. 2025 Jun 30;16:1600233. doi: 10.3389/fimmu.2025.1600233. eCollection 2025. Front Immunol. 2025. PMID: 40661943 Free PMC article. Review.
-
Targeting the Hippo Pathway in Breast Cancer: A Proteomic Analysis of Yes-Associated Protein Inhibition.Int J Mol Sci. 2025 Apr 22;26(9):3943. doi: 10.3390/ijms26093943. Int J Mol Sci. 2025. PMID: 40362184 Free PMC article.
-
Mechanistic Insights into Drug-Induced Guillain-Barré Syndrome: A Large-Cohort Analysis of the FAERS Database.Pharmaceuticals (Basel). 2025 Mar 29;18(4):498. doi: 10.3390/ph18040498. Pharmaceuticals (Basel). 2025. PMID: 40283935 Free PMC article.
References
-
- Malik R., Kopylov M., Gomez-Llorente Y., Jain R., Johnson R.E., Prakash L., Prakash S., Ubarretxena-Belandia I., Aggarwal A.K. Structure and mechanism of B-family DNA polymerase zeta specialized for translesion DNA synthesis. Nat. Struct. Mol. Biol. 2020;27:913–924. doi: 10.1038/s41594-020-0476-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

